Want to download this as a PDF? Download now
Graham Speight, Ezam Uddin, Aysel Heckel, Venu Pullabhatla, James Reid and Lyudmila Georgieva
A wide variety of chromosomal abnormalities are associated with Chronic Lymphocytic Leukaemia (CLL). Currently, comprehensive genetic research into CLL requires multiple testing strategies with high associated costs.
Somatic mutations (Single Nucleotide Variants (SNVs) and insertion/deletions (indels)) can be identified by next generation sequencing (NGS), but copy number alterations (CNAs) currently require additional cytogenetic methods including karyotyping, fluorescence in situ hybridisation (FISH) and microarrays. For a more complete picture, follow-up assays are required to determine the trigger(s) behind the malignant transformation and to characterise the genetic profile to aid research into prognosis and disease management.
In this study, we tested the capability of SureSeq™ NGS panel to overcome the challenges with detecting CNAs currently experienced and provide a possible future single test to be developed for CLL.
The SureSeq hybridisation-based approach was used throughout this study; the workflow of this is outlined in Figure 1.
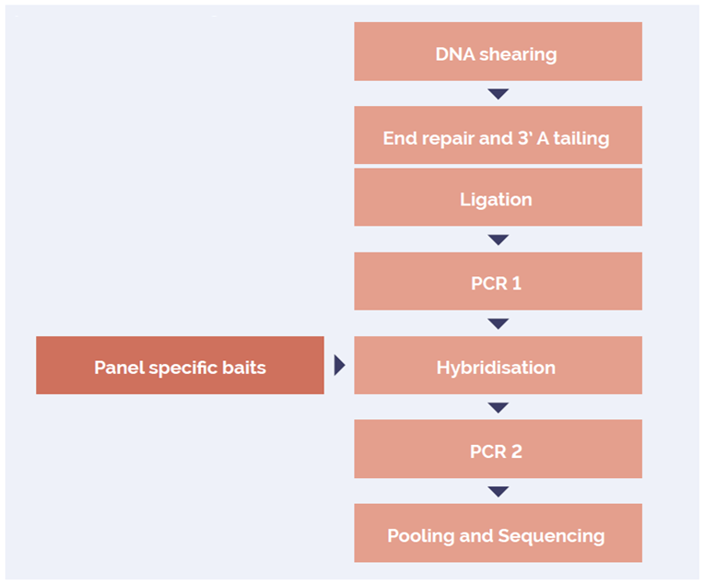 Figure 1: OGT SureSeq workflow. The SureSeq workflow allows users to go from extracted DNA to sequencer in 1.5 days with minimal handling time.
Figure 1: OGT SureSeq workflow. The SureSeq workflow allows users to go from extracted DNA to sequencer in 1.5 days with minimal handling time.
We utilised a SureSeq CLL CNV - 14 gene panel and associated library preparation kit to determine whether this approach can be used for detection of somatic CNAs as well as SNVs and indels.
CLL CNV - 14 gene panel can be used for:
We used a hybridisation-based enrichment approach for library preparation and analysed 15 research samples* with known CNAs. We also analysed 24 samples (Coriell, Camden, NJ) with no known CNVs in the regions of interest which were used as control/reference samples. The resulting libraries were sequenced using a 2x150 bp read length protocol on an Illumina MiSeq®. All research samples were also processed on the Cytoscan™ HD microarray and associated software (Affymetrix®) allowing for comparison of findings of copy number alterations in each sample.
Data sequencing analysis including CNA detection was performed using Interpret, OGT’s complimentary gene variants and CNV detection software.
We achieved high depth (>2000x) and excellent uniformity of coverage across the targeted 14 genes which enabled the confident detection of low frequency gene specific SNVs and indels.
 Figure 2: Example of SF3B1 exon 15 hotspot variant Lys700Glu with frequency 4.8%. Data generated with OGT SureSeq protocol averaging ~2000x deduplicated coverage. Depth of coverage per base (grey).
Figure 2: Example of SF3B1 exon 15 hotspot variant Lys700Glu with frequency 4.8%. Data generated with OGT SureSeq protocol averaging ~2000x deduplicated coverage. Depth of coverage per base (grey).
 Figure 3: Example of TP53 exon 4 frameshift deletion (TP53 c.124del) with frequency 38.9%. Data generated with OGT SureSeq protocol averaging ~2000x deduplicated coverage. Depth of coverage per base (grey).
Figure 3: Example of TP53 exon 4 frameshift deletion (TP53 c.124del) with frequency 38.9%. Data generated with OGT SureSeq protocol averaging ~2000x deduplicated coverage. Depth of coverage per base (grey).
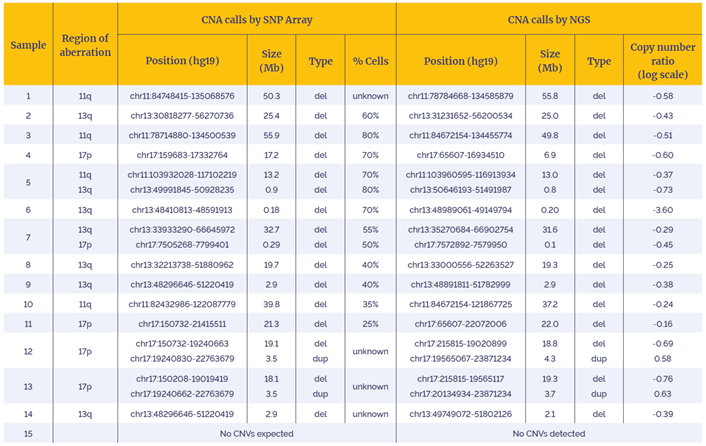 Table 1: Data generated with the CLL CNV - 14 gene panel using a combination of OGT workflow and enhanced CNV detection software was 100% concordant with independent findings (West Midlands Regional Genetics Laboratory – Birmingham, UK).
Table 1: Data generated with the CLL CNV - 14 gene panel using a combination of OGT workflow and enhanced CNV detection software was 100% concordant with independent findings (West Midlands Regional Genetics Laboratory – Birmingham, UK).
Using the OGT workflow we were able to reliably detect somatic copy number alterations in 15 research samples. For all samples, predicted CNAs were found to be 100% concordant with the reported events with the array data.
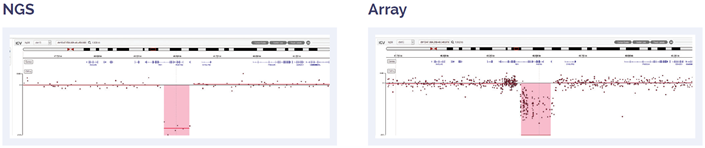 Figure 4: 181kb biallelic deletion within 13q14.2 including RB1.
Figure 4: 181kb biallelic deletion within 13q14.2 including RB1.
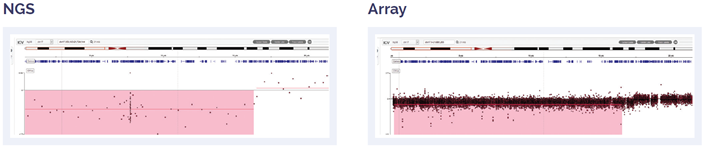 Figure 5: 17.2 Mb deletion of 17pter to p11.2.
Figure 5: 17.2 Mb deletion of 17pter to p11.2.
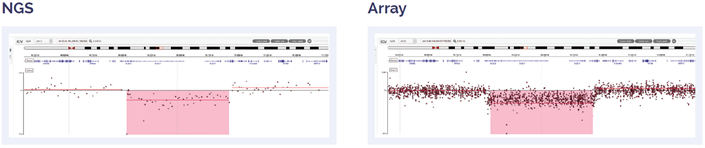 Figure 6: 11 Mb deletion of 13q14.2q14.3, including DLEU2 and DLEU7 genes.
Figure 6: 11 Mb deletion of 13q14.2q14.3, including DLEU2 and DLEU7 genes.
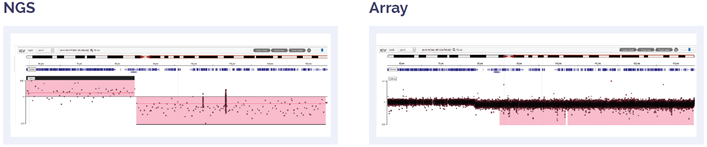 Figure 7: 56 Mb deletion of 11q14.1 to 11q25.
Figure 7: 56 Mb deletion of 11q14.1 to 11q25.
*Samples kindly provided by West Midlands Regional Genetics Laboratory - Birmingham, UK.
SureSeq™: For Research Use Only; Not for Diagnostic Procedures.
