Want to download this as a PDF? Download now
 In the past decade, the advent of next-generation sequencing (NGS) has revolutionised the world of genetic research and is now beginning to impact on clinical testing. The introduction of targeted sequencing – a technique that specifically focuses on sequencing regions associated with causal mutations – has improved speed and accuracy of genetic analysis.
In the past decade, the advent of next-generation sequencing (NGS) has revolutionised the world of genetic research and is now beginning to impact on clinical testing. The introduction of targeted sequencing – a technique that specifically focuses on sequencing regions associated with causal mutations – has improved speed and accuracy of genetic analysis.
One of the centres where NGS has been introduced alongside conventional tools for genetic analysis is the West Midlands Regional Genetics Laboratory (WMRGL) of the Birmingham Women’s NHS Foundation Trust in the UK. It is the largest genetic laboratory in the UK, handling over 50,000 samples per year. The WMRGL team includes Dr Anna Skowronska (R&D scientist), Dr Jane Bryon (MPN team lead) and Ms Joanne Mason (Haemato- Oncology team lead). In this article, they discuss the validation and introduction of a 25-gene myeloid disorder hybridisation-based NGS enrichment panel, recently launched by OGT.
Myeloproliferative neoplasms (MPNs) are a group of conditions affecting the bone marrow which result in an overproduction of blood cells. The three classic types of MPN are all BCR-ABL1 negative and include polycythaemia vera (PV), essential thrombocythaemia (ET) and primary myelofibrosis (PMF). Although there is no current consensus on whether MPNs should be classed as malignant neoplasms, MPNs can result in life-changing symptoms. Without cure, they can progress to more advanced disease and transform to acute myeloid leukaemia – highlighting the need to further understand the molecular mechanisms of disease. Mutations in three genes in particular seem to play a pivotal role in the onset of PV, ET and PMF. Jane explains: “Pathogenic mutations within JAK2, CALR and MPL are consistent with the identification of MPN according to guidelines set out by the World Health Organisation (WHO) and the British Committee for Standards in Haematology.”
One of these mutations, the JAK2 V617F, is very common in all three of the aforementioned forms of MPN, occurring in 97% of cases of PV and in 50-65% of cases of ET and MF1. This specificity makes the JAK2 gene an interesting target for molecular analysis. However, as Jane points out; “There are many samples which do not have mutations in these most commonly involved genes, where a more comprehensive analysis is required, which involves sequencing multiple genes. In addition, investigating a wider range of genes is justified as many samples may have more than one mutation. Variation in progress and outcomes for patients with MPN may reflect the synergistic effects of the different combinations of mutations.”
At WMRGL, digital droplet PCR (ddPCR) is used to identify and quantify JAK2 V617F. In samples negative for this mutation, a sequential analysis regimen based on such assays as Sanger sequencing and capillary electrophoresis was necessary to identify other causative mutations. As Anna explains “although fast, the problem with ddPCR is that it covers only one known mutation per assay and has limited possibilities for multiplexing. With Sanger sequencing, our biggest limitation is sensitivity, which is restricted to 20% allele frequency (AF). This isn’t low enough to detect mutations in all cases as lower allele frequencies are common.”
Jo adds: “Sanger sequencing can be relatively cheap if we focus on one hotspot, however if you want to sequence multiple targets (exons, genes or even exomes), then it actually becomes expensive and laborious. With many potential genes responsible for the underlying pathology, we needed one assay that would detect as many causative mutations as possible in one go, and with good sensitivity.” Jane continues “The main reasons we wanted to implement NGS were to reduce turnaround time, improve sensitivity and get more comprehensive molecular mutation profiling in one assay. And what we’re particularly interested in when we’re looking at NGS panels is that it not only detects the mutations for commonly affected genes like JAK2, CALR and MPL, but also for other genes, which have a role in MPN classification.”
In the fast-moving field of cancer research and diagnostics, being informed and staying up-to-date is a key element in choosing an NGS panel. OGT’s SureSeq™ Myeloid Panel was selected for adoption by WMRGL due to its gene content which was created in collaboration with recognised cancer experts. Designed to save both time and cost, the panel can replace multiple single-gene assays with its comprehensive content of 25 genes implicated in a variety of MPNs – including all of the genes recommended by the WHO and the British Committee for Standards in Haematology. As Anna simply puts, “OGT’s SureSeq Myeloid Panel contains all the genes we want and more.” (Table 1).
 Table 1: The 25 genes that make up OGT’s SureSeq Myeloid Panel are all implicated in different forms of MPN.
Table 1: The 25 genes that make up OGT’s SureSeq Myeloid Panel are all implicated in different forms of MPN.
OGT’s Myeloid Panel was assessed at the WMRGL by analysing 160 samples including 30 known mutations, where a mutation had previously been detected by ddPCR or Sanger sequencing. “We’ve found that the SureSeq Myeloid Panel performs very well and has detected all known positives with an excellent variant detection of around 1%,” Anna reports. “Working with OGT has been very positive, we’ve been happy to collaborate with them and this has made it easy to customise their products.”
Overview of the 30 mutations detected by OGT’s SureSeq panel as part of the validation process:
Highlighting the success of the SureSeq Myeloid Panel over conventional gene analysis techniques, Jane and Anna present two particular case studies that demonstrate the sensitive and comprehensive analysis that can be performed using NGS.
Anna highlights the importance of reliability in the detection of mutations by demonstrating a case where a mutation was missed by conventional genetic analysis. It involved the analysis of exon 14 of the JAK2 gene by ddPCR to detect the common V617F mutation.
 Whereas ddPCR is normally well suited to detect this variant, this case was complicated by the presence of a rare second mutation only three bases upstream (c.1852C>T). “The V617F mutation was not detected because of this second variant under the probe binding site, resulting in a false negative result,” explains Anna. When the same sample was subsequently re-analysed by NGS using the SureSeq Myeloid Panel, the V617F and the c.1852C>T were both detected at AF 32% (Figure 1), and the sequencing data confirmed that both mutations were on the same allele (in cis). Anna comments, “this may be a rare event, but with NGS we may actually discover more of such incidents that could not be picked up otherwise.”
Whereas ddPCR is normally well suited to detect this variant, this case was complicated by the presence of a rare second mutation only three bases upstream (c.1852C>T). “The V617F mutation was not detected because of this second variant under the probe binding site, resulting in a false negative result,” explains Anna. When the same sample was subsequently re-analysed by NGS using the SureSeq Myeloid Panel, the V617F and the c.1852C>T were both detected at AF 32% (Figure 1), and the sequencing data confirmed that both mutations were on the same allele (in cis). Anna comments, “this may be a rare event, but with NGS we may actually discover more of such incidents that could not be picked up otherwise.”
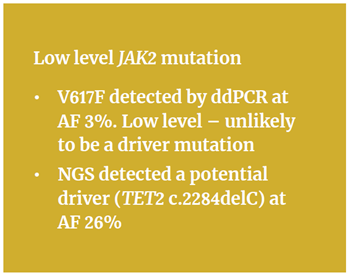 A good example of the need for comprehensive, multi-gene tests to fully understand the genetic basis of MPN on a case-by-case basis was a sample analysed with ddPCR, discovered to have the V617F mutation (AF 3%). However, “because it was found to be a low level mutation, this raised doubts regarding the underlying genetic causes,” Anna explains.
A good example of the need for comprehensive, multi-gene tests to fully understand the genetic basis of MPN on a case-by-case basis was a sample analysed with ddPCR, discovered to have the V617F mutation (AF 3%). However, “because it was found to be a low level mutation, this raised doubts regarding the underlying genetic causes,” Anna explains.
“We believed a low level JAK2 mutation was unlikely be the driver mutation,” she elaborates. “This is why we then re-analysed the sample using the SureSeq Myeloid Panel to investigate all of the 25 genes.” This more comprehensive analysis revealed a mutation in the TET2 gene – a gene that is not yet routinely analysed – which was far more likely to be a driver mutation for MPN. The frameshift mutation (c.2284delC p.(His762ThrfsTer51)) was reported by NGS at AF 26% (Figure 2).
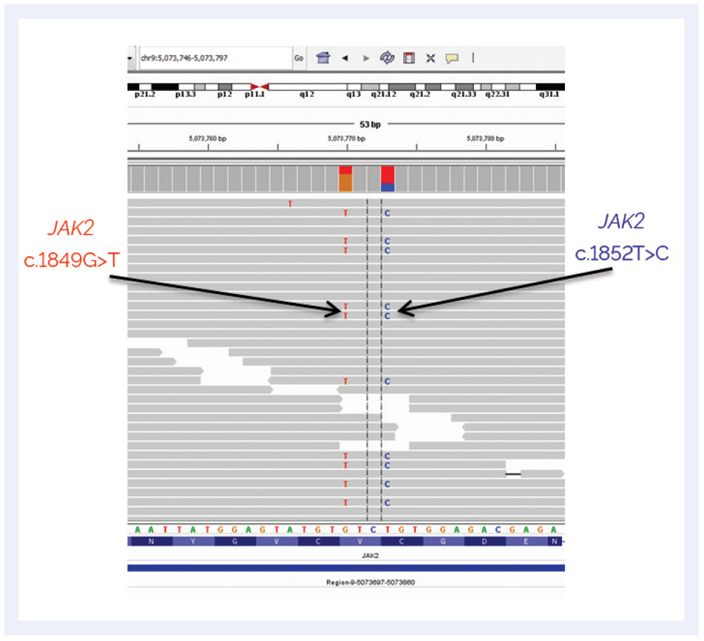 Figure 1: IGV genome viewer showing the analysis of the NGS data with arrows showing the newly identified mutations not detected by ddPCR.
Figure 1: IGV genome viewer showing the analysis of the NGS data with arrows showing the newly identified mutations not detected by ddPCR.
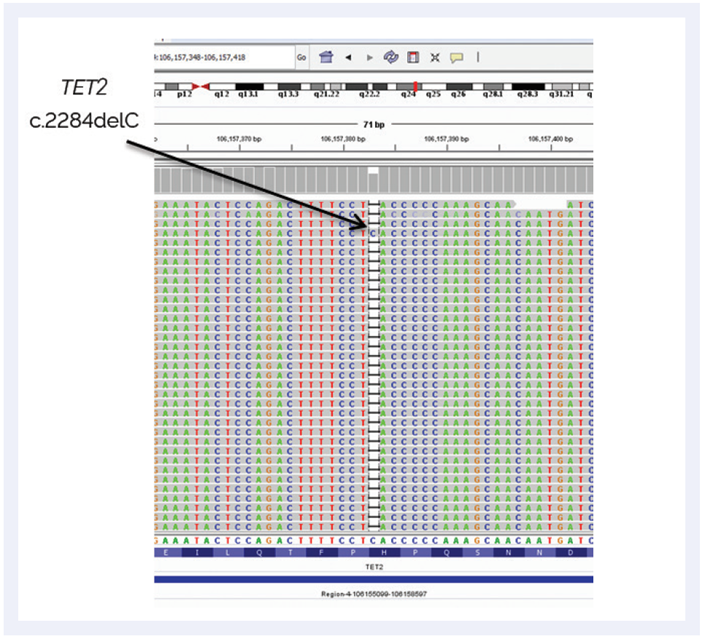 Figure 2: NGS analysis showing the TET2 mutation, which is a potential driver for MPN.
Figure 2: NGS analysis showing the TET2 mutation, which is a potential driver for MPN.
TET2 mutations have been reported2 to occur in a range of myeloproliferative disorders, suggesting that disruption of this gene can be an early event in the development of the disease. Therefore, the availability of NGS panels such as the 25-gene SureSeq panel offers researchers the opportunity to analyse for these genes in a straightforward manner. “It is clear that it is important to carry out comprehensive genetic analysis,” says Anna, “and this panel offers this possibility in one assay.”
MPN cases with low levels of JAK2 mutations are frequently detected in routine analysis. In such events the JAK2 mutation may represent a sub-clone, and other gene mutations that define the dominant clone are expected to be present. With the increasing availability of targeted therapies it will be important to understand the complete mutation profile of each patient.
Next-generation sequencing is clearly continuing to revolutionise our understanding of genetics. The introduction of multi-gene, targeted NGS panels such as OGT’s SureSeq Myeloid Panel brings a range of benefits to scientists carrying out research into debilitating diseases. For the scientists at the WMRGL, incorporating NGS into their existing workflow has been very smooth. Anna notes: “The library enrichment protocol is very well optimised and easy to follow, we were positively surprised right from the very first analysis by the level of mutation detection achievable. In addition, the OGT team has been very helpful and accessible when we needed any kind of technical support.”
The study performed by the WMRGL and in particular the two highlighted case studies exemplify the way in which the SureSeq panel has allowed scientists at the WMRGL to identify important mutations. This includes mutations which had been overlooked by conventional techniques, clearly demonstrating its value in developing a better understanding of myeloproliferative disorders.
Looking forward, WMRGL team expects an expanding role for the SureSeq Myeloid Panel. “As a result of the excellent results, ease of use and accessible customer support from OGT, we have already implemented the SureSeq Myeloid Panel into routine use. We are moving towards wider implementation of NGS technologies. This seems to be the trend for the near future.”
SureSeq: For Research Use Only; Not for Diagnostic Procedures
