Want to download this as a PDF? Download now
Lyudmila Georgieva, Stephanie Carpenter, Natalie Milner, Venu Pullabhatla, James Reid, Kay Wiebrands, Aysel Heckel, Graham Speight
Loss-of-function mutations in tumour suppressor genes BRCA1 and BRCA2 have been implicated in an increased risk for breast and ovarian cancer1,2. Screening for germline mutations in these genes allows research into familial risk of developing breast and ovarian cancer. In addition, assessment of somatic mutations in tumour samples can help research into tumour development, drug response and the development of new therapies.
A wide range of genetic variations are associated with breast and ovarian cancer, including single nucleotide variants (SNVs), small insertions/deletions (indels) and copy-number variations (CNVs). For more than a decade, the gold standard for mutational screening has been Sanger sequencing and multiplex ligation-dependent probe amplification (MLPA), imposing significant time and cost burden.
Advances in next-generation sequencing (NGS) now allows for the reliable detection of CNVs in addition to SNVs/indels in a single assay.
In this study, we tested the capability of the SureSeq™ Breast Cancer + CNV Panel to overcome the challenges currently experienced and provide a possible future single assay to be developed for breast and ovarian cancer.
The SureSeq hybridisation-based approach was used to assess the detection of CNVs as well as SNVs and indels in a single assay; the workflow is outlined in Figure 1.
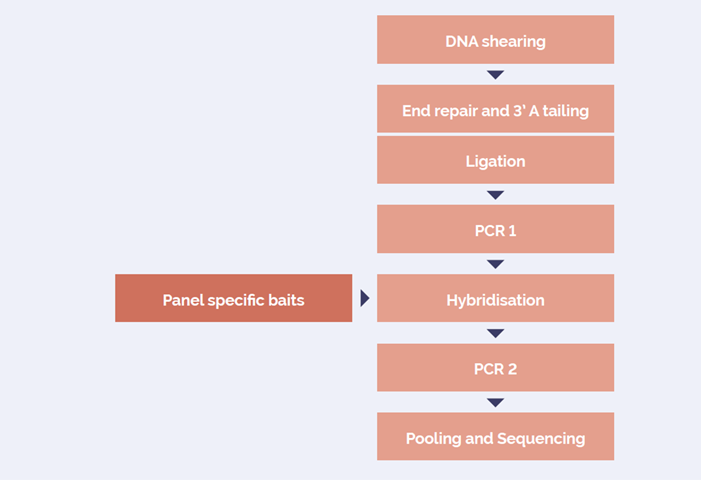
Figure 1: OGT SureSeq workflow. The SureSeq workflow allows users to go from extracted DNA to sequencer in 1.5 days with minimal handling time.
The SureSeq Breast Cancer + CNV Panel can be used for detection of SNV/indels and CNVs in 7 genes - ATM, BRCA1, BRCA2, TP53, CHEK2, PALB2, PTEN.
A number of different sample types were profiled:
The resulting libraries were sequenced using a 2x150 bp read length (v2) protocol on an Illumina MiSeq®.
CNV detection concordance was assessed by comparing NGS calls to events reported by an orthogonal technology, MLPA for set 1 samples and CytoSure® Cancer +SNP Arrays and CytoSure software (OGT) for set 2 samples.
Data sequencing analysis including CNV detection was performed using Interpret, OGT’s complimentary NGS analysis software.
Facilitated by OGT’s expert bait design, the hybridisation-based SureSeq Breast Cancer + CNV Panel delivers excellent coverage uniformity, allowing consistent detection of SNVs and indels in germline samples, as well as somatic analysis on FPPE tissues down to 1% VAF (Figures 2 and 3).
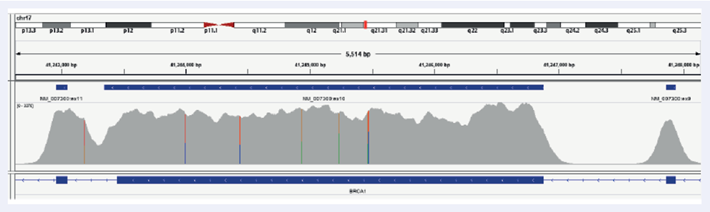
Figure 2: Illustration of the excellent coverage uniformity of BRCA1 exons 9, 10 and 11 in FFPE samples. Depth of coverage per base (grey). Gene coding region as defined by RefSeq (bottom track).
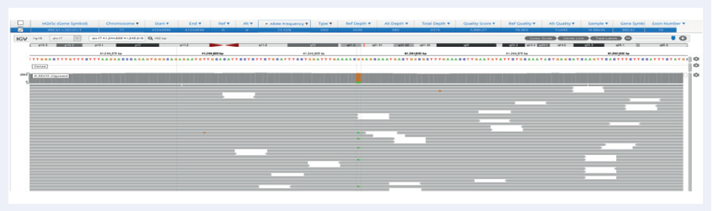
Figure 3: Excellent uniformity and high depth of coverage allowing confident detection of variants. Example of BRCA1 exon 10 missense variant Pro871Leu (rs799917) with frequency 15.9% identified in FFPE samples. Data generated with OGT SureSeq protocol averaging ~2000x deduplicated coverage. Depth of coverage per base (grey).
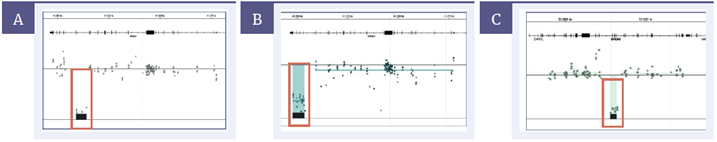
Figure 4: Detection of germline CNVs in BRCA1 and BRCA2. [A] Deletion of ex20 BRCA1 (4.99kb), [B] deletion of ex21-23 BRCA1 (5.93kb) [C] Deletion of ex14-17 BRCA2 (4.21kb)
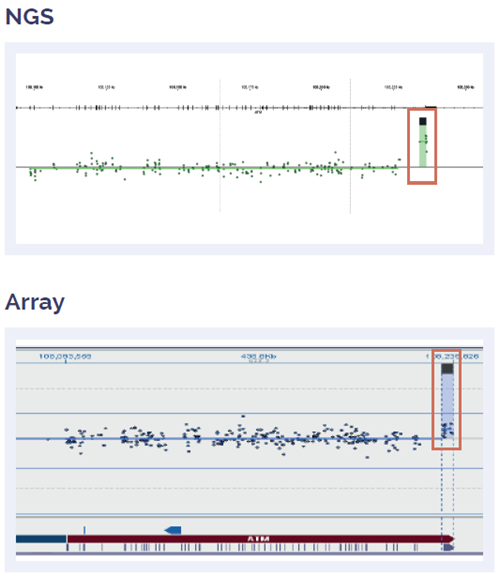
Figure 5: Somatic ATM duplication of exon 62-63, fully concordant with array data.
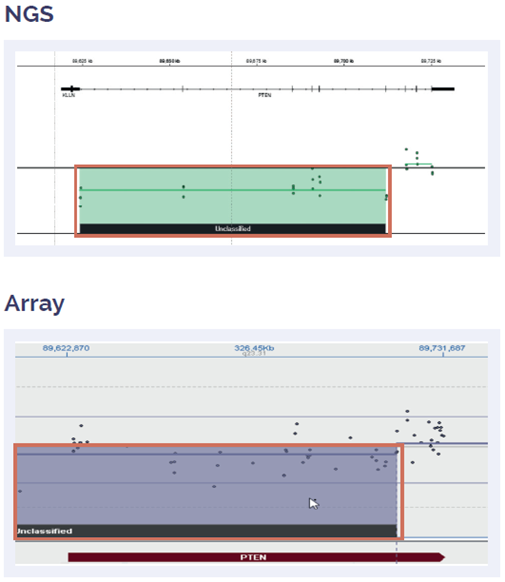
Figure 6: Somatic PTEN deletion of exon 1-5, fully concordant with array data
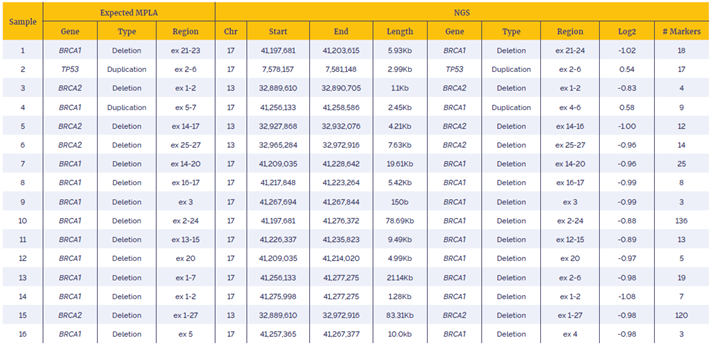
Table 1: Data generated with the SureSeq Breast Cancer + CNV Panel using a combination of OGT workflow and Interpret software showed very high concordance with alternative technology (MLPA). NGS data annotations are is based on NM_007300 (BRCA1), NM_000546 (TP53) and NM_000059 (BRCA2)
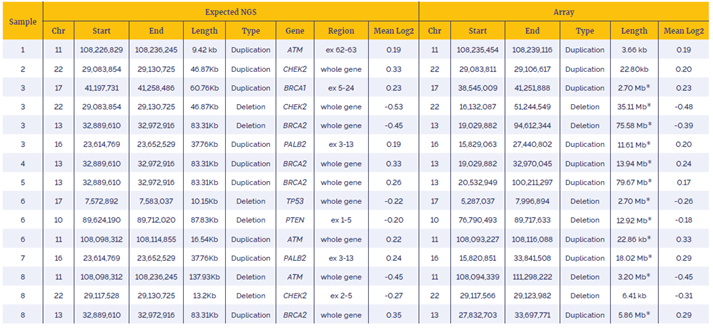
Table 2: Data generated with the SureSeq Breast Cancer + CNV Panel using a combination of OGT workflow and enhanced CNV detection software shows very high concordant with alternative technology (OGT array).
SureSeq and CytoSure: For research use only. Not for diagnostic procedures
