Want to download this as a PDF? Download now

Investigation of exon-level resolution microarray technology for the identification of important mutations in clinical genetics research.
A vital aspect of clinical genetics research is the accurate and reliable detection of genetic aberrations, such as copy number variation (CNV) and loss of heterozygosity (LOH), particularly in the study of developmental disorders. Next generation sequencing (NGS) technology struggles to detect structural aberrations reliably and, as such, array comparative genomic hybridisation (aCGH) remains the gold standard for CNV detection1.
The use of microarray technology, however, is only as effective as the pace it keeps with ongoing discoveries in genetics. Content needs to be up to date to ensure the continued targeting of the most disease-relevant regions. The majority of commercially available arrays offer gene-level resolution, enabling the identification of CNVs containing whole gene or multiple exon variation. However, this level of resolution is often insufficient, as it does not allow researchers to associate different phenotypes with smaller (exonic) aberrations within a given gene. To do this, exon-level resolution is required, which has traditionally necessitated the use of custom ultra-high resolution arrays at greater expense. The ability to use exonlevel resolution routinely would therefore be a real advantage in clinical genetics research.
We conducted a comparative study of a number of array platforms. These included the OGT CytoSure® Constitutional v3 8x60k (containing up to 60,000 60-mer probes), Platform I 8x850k (containing up to 850,000 50-mer probes), and Platform A 750k (containing up to 750,000 25- mer probes) arrays, which were assessed for detection rate, ease of analysis, throughput and cost.
From our initial assessment, it was clear that OGT understood the need for the latest data and exon-level resolution in the development of its CytoSure Constitutional v3 array. The probe design had been informed by new data from the recent Deciphering Developmental Disorders (DDD) project2, as well as ClinGen: A National Institutes of Health (NIH) funded resource dedicated to defining the relevance of genes and variants for use in precision medicine and research (formerly known as ISCA/ICCG)3.
The DDD project aimed to advance the understanding of developmental disorders using multiple technologies, including high-density aCGH (using 2 million probes to interrogate each sample, with each exon covered by at least 5 probes), whole exome sequencing (WES), and whole genome sequencing (WGS). The project has led to huge developments in the DECIPHER database (a vast resource of curated phenotypic and genotypic data), delivering data from 12,000 trio exomes, and discovering at least 12 new genes involved in developmental disorders. As a result, we have access to a new, very evidence-based, list of over 1,400 genes associated with developmental disorders. Any new array platform would greatly benefit from being informed by this list, and currently the CytoSure Constitutional v3 is the only commercially available array based on this latest data.
 Unlike the other arrays studied, the OGT platform also provides exon-level resolution, with a highly targeted design to maximise usefulness (Figure 1). In addition to backbone coverage throughout the genome, the targeted 60-mer probes are placed at higher density across exons of the latest research-validated disease-associated genes. This enables high-resolution single exon CNV detection and the discovery of de novo variants in a cost-effective manner.
Unlike the other arrays studied, the OGT platform also provides exon-level resolution, with a highly targeted design to maximise usefulness (Figure 1). In addition to backbone coverage throughout the genome, the targeted 60-mer probes are placed at higher density across exons of the latest research-validated disease-associated genes. This enables high-resolution single exon CNV detection and the discovery of de novo variants in a cost-effective manner.
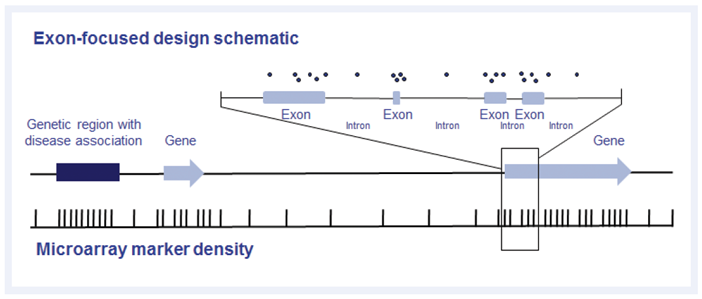 Figure 1: Example of exon-focused design, as used on the CytoSure Constitutional v3 array. In addition to an evenly spaced backbone, probes are targeted to genes and non-genic regions associated with developmental disorders using the latest data from the Deciphering Developmental Disorders study2 and ClinGen3.
Figure 1: Example of exon-focused design, as used on the CytoSure Constitutional v3 array. In addition to an evenly spaced backbone, probes are targeted to genes and non-genic regions associated with developmental disorders using the latest data from the Deciphering Developmental Disorders study2 and ClinGen3.
The WMRGL has a bank of thousands of abnormal samples, of which several with aberrations known to be difficult to identify and interpret were selected for the study.
Testing the performance and consistency of the probes, we examined a sample with a large 8.7Mb deletion in the region known to be associated with Chromosome 1q41-q42 deletion syndrome, on the CytoSure Constitutional v3 8x60k array (Figure 2). An average log2 ratio of -0.9 with all 119 probes below the set threshold of -0.6 indicated that the probe behaviour is highly consistent.
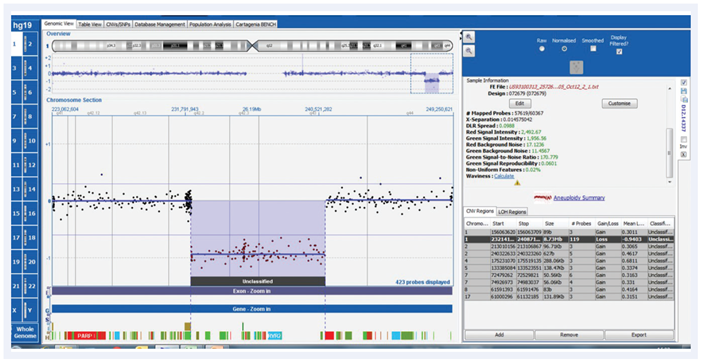 Figure 2: CytoSure Constitutional v3 array probes across 8.7Mb 1q41-q42 deletion displayed using CytoSure Interpret Software. An average log2 ratio of -0.9, with all 119 probes on the deletion below a threshold of -0.6 indicates highly consistent probe behaviour.
Figure 2: CytoSure Constitutional v3 array probes across 8.7Mb 1q41-q42 deletion displayed using CytoSure Interpret Software. An average log2 ratio of -0.9, with all 119 probes on the deletion below a threshold of -0.6 indicates highly consistent probe behaviour.
We also tested the CytoSure Constitutional v3 for calling an aberration in MEF2C, an important gene not present on the original ClinGen gene list (Figure 3). MEF2C is a transcriptional activator that specifically binds MEF2, an element present in the regulatory regions of many muscle-specific genes4 . Microdeletions are associated with severe developmental delay, hypotonia and seizures. Previous study of this sample required manual calling of the aberration due to the presence of only two probes, necessitating a higher resolution array to confirm. On the CytoSure Constitutional v3 array, 36 probes are mapped across multiple exons enabling robust detection of this MEF2C deletion.
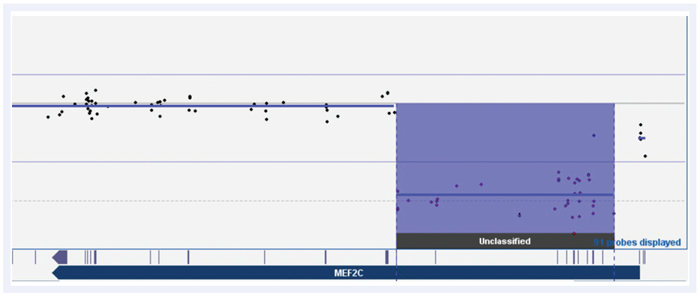 Figure 3: CytoSure Constitutional v3 array probes across the MEF2C gene displayed using CytoSure Interpret Software. Deletion with a minimum size of 68kb called by 36 probes across multiple exons, which had previously required manual calling and validation from two probes on our previous array platform.
Figure 3: CytoSure Constitutional v3 array probes across the MEF2C gene displayed using CytoSure Interpret Software. Deletion with a minimum size of 68kb called by 36 probes across multiple exons, which had previously required manual calling and validation from two probes on our previous array platform.
Another challenging sample was that of calling an intragenic duplication of a single exon in MID1, which is associated with Opitz-G syndrome5 (Figure 4). In our previous investigation, this required multiple attempts with different technologies over a seven-year period, including sequence analysis followed by three different arrays, before finally confirming on a custom 2x105k OGT CytoSure Chromosome X array. Using the CytoSure Constitutional v3 array, we found there were four probe calls within an interval of a minimum size of 500bp, indicating that with the CytoSure Constitutional v3 array we could have detected this disease-associated finding years previously.
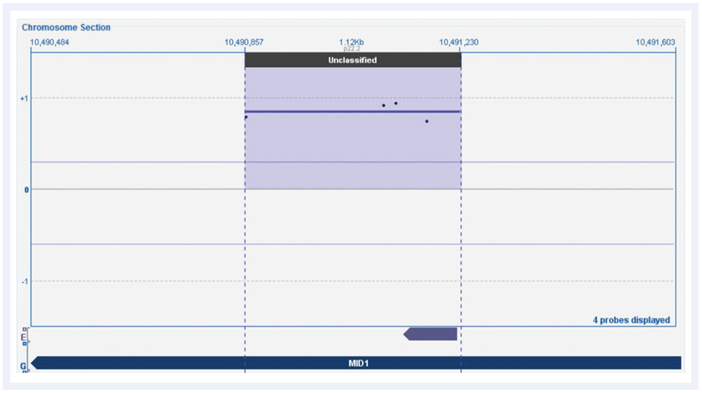 Figure 4: CytoSure Constitutional v3 array probes across MID1 gene displayed using CytoSure Interpret Software. The log2 ratio trace identifies a single exon duplication called by four probes, which had previously required several attempts over seven years to call and validate.
Figure 4: CytoSure Constitutional v3 array probes across MID1 gene displayed using CytoSure Interpret Software. The log2 ratio trace identifies a single exon duplication called by four probes, which had previously required several attempts over seven years to call and validate.

Powerful software is required to interpret array data, especially that from difficult samples. Mosaic trisomy 18 is an example of a problematic aberration to identify simply from a copy number trace. This makes feature-rich, but easy-to-use, software, such as OGT’s CytoSure Interpret Software novel as it can help eliminate subjective assessment. We examined a mosaic trisomy 18 sample with the CytoSure Constitutional v3 array (Figure 5).
From the standard copy number trace, the upwards movement of the probes at chromosome 18 is unclear. However, OGT’s CytoSure Interpret Software provides an aneuploidy summary plot — a box and whisker plot that displays the log2 ratios of every individual probe, showing the mean and median. The value of the aneuploidy summary plot is immediately clear. There is a noticeable rise for chromosome 18, relative to the others, which would be difficult to identify otherwise.
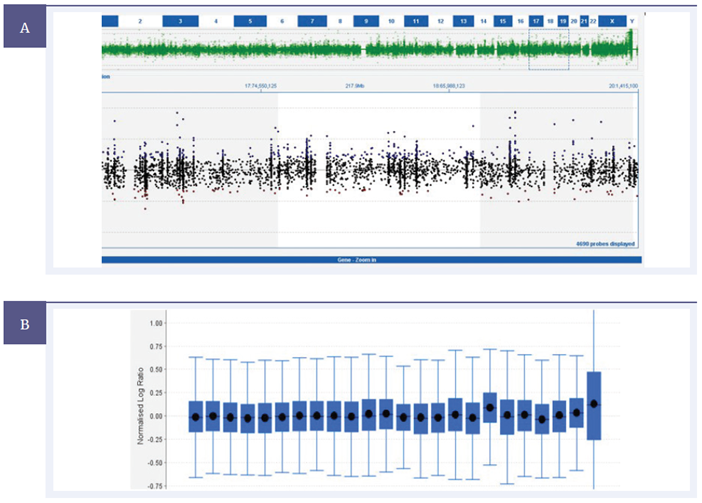 Figure 5: Copy number trace of CytoSure Constitutional v3 probes across Chromosome 18 from a mosaic trisomy 18 sample using the CytoSure Interpret Software. (A) Mosaic trisomy 18 is readily identifiable due to ease of interpretation of the aneuploidy summary plot (B) a function provided in CytoSure Interpret Software.
Figure 5: Copy number trace of CytoSure Constitutional v3 probes across Chromosome 18 from a mosaic trisomy 18 sample using the CytoSure Interpret Software. (A) Mosaic trisomy 18 is readily identifiable due to ease of interpretation of the aneuploidy summary plot (B) a function provided in CytoSure Interpret Software.
Comparing the three platforms falling within our study, we took samples containing difficult to detect aberrations and determined which of the platforms would have detected the aberrations.
The first sample contained a small deletion in HDAC8, which has an important role in tagging epigenetic regulation in early development6 . The CytoSure Constitutional v3 8x60k array was able to identify a 1.17kb deletion (minimum) across two exons from seven probes (Figure 6a). To evaluate the likely performance of the other arrays, probe positions in this region for all three platforms were plotted on the University California, Santa Cruz (UCSC) gene browser, enabling a predictive comparison (Figure 6b). Based on the probe locations, it is evident that the OGT platform alone would detect this aberration to the exon level, including both minimum and maximum sizes. Platform I typically requires around 5–10 probes to make a call, with just three probes present in this region. Platform A (utilising shorter, 25-mer probes) requires at least 10–20 probes to call, with only two present. Neither would have detected the aberration, even if the deletion had spanned the entire maximum region.
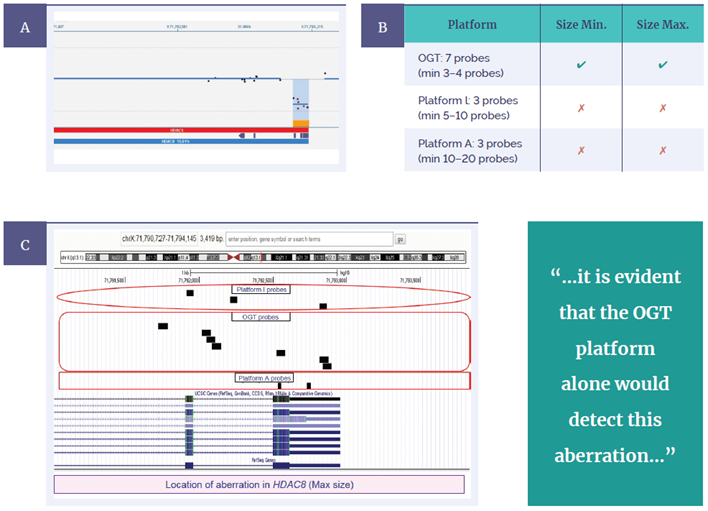 Figure 6: CytoSure Constitutional v3 probes across the HDAC8 gene displayed using CytoSure Interpret Software. (A) A 1.17kb deletion (minimum) is called across two exons from seven probes. OGT probe design provides superior detection, evidenced by being the only platform able to call the aberration to exon level, including both minimum and maximum sizes (B) Predictive comparison of probe locations for OGT, Platform I and platform A, using BED files on the UCSC genome browser (C).
Figure 6: CytoSure Constitutional v3 probes across the HDAC8 gene displayed using CytoSure Interpret Software. (A) A 1.17kb deletion (minimum) is called across two exons from seven probes. OGT probe design provides superior detection, evidenced by being the only platform able to call the aberration to exon level, including both minimum and maximum sizes (B) Predictive comparison of probe locations for OGT, Platform I and platform A, using BED files on the UCSC genome browser (C).
A second sample contained an exonic deletion in MED13L, a gene with an important role in early transcription in the brain and heart7. Repeating the test using this sample, we confirmed a 269bp deletion (minimum) across one exon called by the CytoSure Constitutional v3 8x60K array from three probes (Figure 7a). The predictive comparison indicated that the OGT platform would again be the only one to call the aberration, with both minimum and maximum sizes (Figure 7b). In this case, three probes represented Platform I across the maximum size determined, while Platform A had four probes present, but none on the exon itself. Given these platforms require many more probes to make a call, neither would have detected this imbalance.
These aberrations were both originally detected using ultra high-resolution aCGH carried out as a part of the DDD study. In both cases, the gene level coverage of Platform I and Platform A would prevent the detection of these aberrations, in spite of the higher total number of probes used by each platform. Only the OGT CytoSure Constitutional v3 array has the exon-focused probe design as well as the updated DDD and ClinGen probe content required for detection. Ultimately, a researcher has a better chance of identifying relevant variants using this array than with the alternative array platforms.
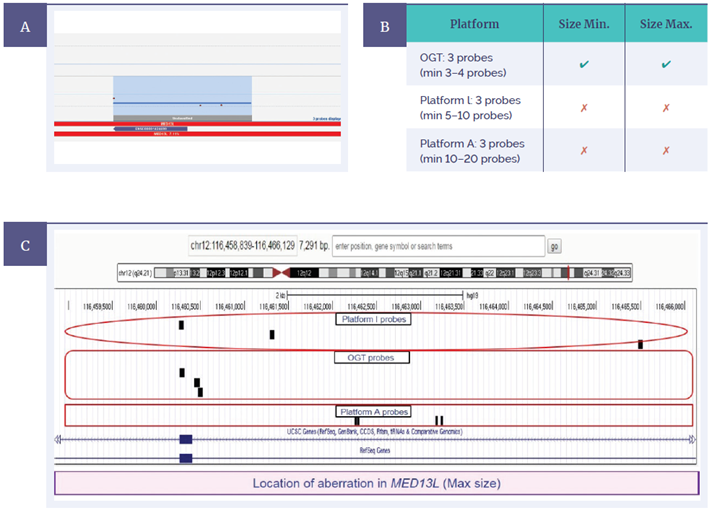 Figure 7: CytoSure Constitutional v3 probes across the MED13L gene displayed using CytoSure Interpret Software (A) A 269bp deletion (minimum) is called across one exon from three probes. OGT probe design provides superior detection, evidenced by being the only platform able to call the aberration to exon level, including both minimum and maximum sizes (B) Predictive comparison of probe locations for OGT, Platform I and Platform A using BED files on UCSC genome browser (C).
Figure 7: CytoSure Constitutional v3 probes across the MED13L gene displayed using CytoSure Interpret Software (A) A 269bp deletion (minimum) is called across one exon from three probes. OGT probe design provides superior detection, evidenced by being the only platform able to call the aberration to exon level, including both minimum and maximum sizes (B) Predictive comparison of probe locations for OGT, Platform I and Platform A using BED files on UCSC genome browser (C).
SNP probe-based arrays can also be a powerful tool for researchers due to the natural variance of SNPs in the genome. They have the capability to detect absence of heterozygosity (AOH) or uniparental disomy (UPD) very accurately. We compared Platform A (a high density array design) with an enhanced version of the OGT platform, the CytoSure Constitutional v3 +LOH 4x180k array, which includes SNP probes.
A sample obtained from an individual with Bartter syndrome was initially analysed on Platform A. Bartter type 3 is associated with an aberration in the gene CLCNKB while type 4B is associated with aberrations in both CLCNKA and CLCNKB8 (which are highly homologous). Platform A software called a deletion across genes CLCNKA and CLCNKB, suggesting both genes were involved.
 Another predictive comparison on the UCSC genome browser presented probe locations from an unfiltered (higher probe density) Platform A array against those of Platform I and the standard CytoSure Constitutional v3 8x60k array (Figure 8a). A small number of probes within CLCNKB, but none within CLCNKA, represented the higher density Platform A. However, both CLCNKA and CLCNKB have good probe coverage on the OGT CytoSure Constitutional v3 8x60K array. It is therefore evident that high density does not equal high resolution.
Another predictive comparison on the UCSC genome browser presented probe locations from an unfiltered (higher probe density) Platform A array against those of Platform I and the standard CytoSure Constitutional v3 8x60k array (Figure 8a). A small number of probes within CLCNKB, but none within CLCNKA, represented the higher density Platform A. However, both CLCNKA and CLCNKB have good probe coverage on the OGT CytoSure Constitutional v3 8x60K array. It is therefore evident that high density does not equal high resolution.
To verify the better coverage on the OGT platform, we ran the same sample on the CytoSure Constitutional v3 +LOH 4x180k array (Figure 8b). This indicated regions of homozygosity consistent with segmented UPD, and a very clear homozygous deletion. Multiple probes were present in both genes, clearly demonstrating no deletion of CLCNKA, and homozygous deletion of CLCNKB. This was consistent with Bartter type 3. These data suggest that the 60-mer probes used by OGT allow better design for regions with high homologies, which in this case enabled the delineation of the two syndrome types in this sample.
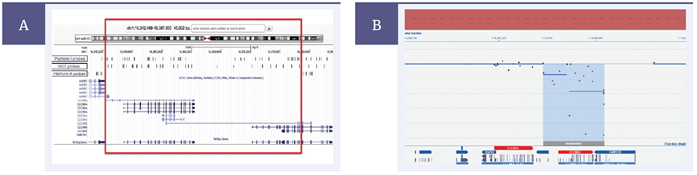 Figure 8: Predictive comparison of probe locations across Bartter syndrome-associated genes, CLCNKA and CLCNKB, for OGT, Platform I and Platform A using BED files on UCSC genome browser. (A) Platform A only contains probes targeted to CLCNKB and therefore cannot conclusively identify genes affected by LOH aberration (red box indicates area without any probe coverage in Platform A). The CytoSure Constitutional v3 +LOH array contains probes across both CLCNKA and CLCNKB genes displayed using CytoSure Interpret Software (B). The CLCNKB gene deletion is readily identified despite >90% homology with CLCNKA.
Figure 8: Predictive comparison of probe locations across Bartter syndrome-associated genes, CLCNKA and CLCNKB, for OGT, Platform I and Platform A using BED files on UCSC genome browser. (A) Platform A only contains probes targeted to CLCNKB and therefore cannot conclusively identify genes affected by LOH aberration (red box indicates area without any probe coverage in Platform A). The CytoSure Constitutional v3 +LOH array contains probes across both CLCNKA and CLCNKB genes displayed using CytoSure Interpret Software (B). The CLCNKB gene deletion is readily identified despite >90% homology with CLCNKA.
Accurate genetic analyses require up-to-date and cost-effective technologies. The work described here demonstrates that the OGT CytoSure Constitutional v3 array, with its exon-focused 60-mer probes, can outperform Platform I and Platform A in certain instances, particularly where exon-level detection is key. In our investigations, we found that, with design informed by the latest data from the DDD study and ClinGen, the OGT platform could detect relevant variants, including single exon deletions, which were not detectable by the other platforms and therefore removed the necessity for additional investigations. In a typical year, we analyse around 5000 samples. From our initial studies, we would anticipate being able to identify disease-relevant aberrations in at least an additional 20–30 samples than would be possible using other available platforms. Supplied with the array is also OGT’s CytoSure Interpret Software, which we found provides rapid and powerful analysis of aCGH data and can be highly customised with additional data-tracks and filters.
We have now implemented the OGT CytoSure Constitutional v3 array platform at the WMRGL for the study of developmental disorders in both pre- (foetal malformation) and postnatal samples. The quality of the evidence base behind the array design allows for very comprehensive analysis of the key exons, genes and regions implicated in all known developmental disorders. We found it provided the claimed exon-level resolution and out-performed the other platforms in the ability to detect exon level variants. In addition, the speed, flexibility and versatility of OGT’s CytoSure Interpret Software was very important in our decision-making.
Further to this, OGT continues to develop the CytoSure array range, responding to our requests and implementing enhancements, such as new software tracks and features, as a matter of course. This support provided by OGT has been first-class and puts us in good stead for the future.

CytoSure®: For Research Use Only; Not for Diagnostic Procedures.
