Want to download this as a PDF? Download now
This application note describes the detailed clinical and analytical studies carried out to meet the performance required to achieve FDA clearance of the eight probes for acute myeloid leukemia (AML) and myelodysplastic syndromes (MDS) listed in Table 1. In addition, we present data demonstrating excellent analytical reproducibility of the probes and extensive stability studies. During the course of these studies, which spanned four sites, worldwide, over 2,500 replicates were run with no technical failures. The rigorous standards required for gaining FDA clearance are recognized globally and emphasize the exceptional quality of the CytoCell® FISH probes from OGT.
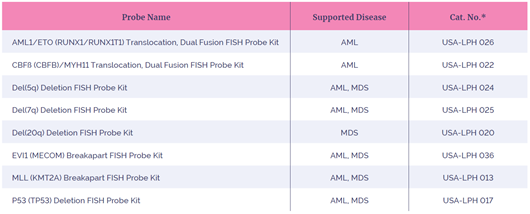 Table 1: The eight FISH probes in this study
Table 1: The eight FISH probes in this study
*Kit includes FISH probe and DAPI
The CytoCell AML/MDS range of FISH probe test kits are fluorescence in situ hybridization (FISH) tests used to detect common chromosomal rearrangements in fixed bone marrow specimens from patients with acute myeloid leukemia (AML) or myelodysplastic syndrome (MDS). The tests are indicated for the characterization of patient specimens consistent with World Health Organization guidelines for Classification of Tumours of Haematopoietic and Lymphoid Tissues (Revised 4th Edition) and in conjunction with other clinicopathological criteria. The assay results are to be interpreted by a qualified pathologist or cytogeneticist. The tests are not intended for use as a stand-alone diagnostic, disease screening, or as a companion diagnostic.
Refer to individual test kit Package Insert for the specific intended use and limitations. For In Vitro Diagnostic Use. Rx only.
AML and MDS are neoplastic hematological disorders that arise from myeloid progenitor cells in the bone marrow. AML is characterized by the clonal expansion of myeloid blasts in the peripheral blood, bone marrow or other tissues, whilst MDS is characterized by the simultaneous proliferation and apoptosis of hemopoietic cells1. According to the World Health Organization (WHO), the global incidence for MDS is 3-5 cases per 100,000 (non-age corrected) with approximately 10,000 new cases of MDS diagnosed annually in the USA1. The Surveillance Epidemiology and End Results (SEER) statistics present a similar picture for AML with a USA incidence of 4.3 per 100,000 (non-sex specific)2.
The National Comprehensive Cancer Network Guidelines in Oncology (NCCN Guidelines®) recognizes that FISH testing is a valuable tool in determining and clarifying the presence of common chromosomal rearrangements in the investigation of AML and MDS. The information from cytogenetic analysis is used for clinical management of patients consistent with WHO guidelines in AML3 and the prognostic scoring systems of MDS4. FISH testing can also be used to monitor patient progress during and after treatment for AML and MDS, by assessing the presence or absence of an abnormality within a bone marrow sample5,6.
An extensive range of studies were conducted to determine the performance characteristics of the CytoCell AML/MDS FISH Probe Kits. All CytoCell FISH probes are designed to give tight, bright signals that are easy-to-score with minimal background, enabling rapid analysis and reduced repeat testing.
An example of probe design and typical results is shown in Figure 1.
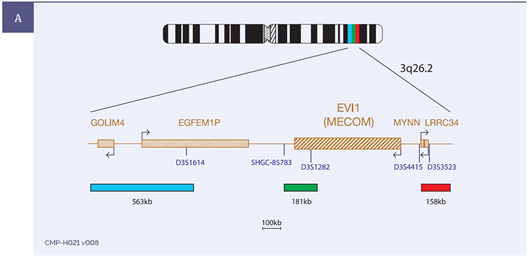
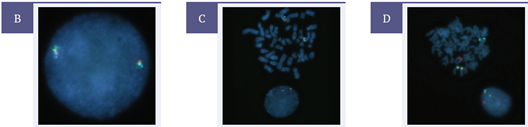 Figure 1: Example of the results obtained for USA-LPH 036: EVI1 (MECOM), including [A] the probe design, [B] expected normal 2RGB, [C] a sample showing an inversion involving the MECOM region 1B/1RG/1RGB and [D] a sample showing a translocation involving the MECOM region 1R/1GB/1RGB.
Figure 1: Example of the results obtained for USA-LPH 036: EVI1 (MECOM), including [A] the probe design, [B] expected normal 2RGB, [C] a sample showing an inversion involving the MECOM region 1B/1RG/1RGB and [D] a sample showing a translocation involving the MECOM region 1R/1GB/1RGB.
Analytical specificity can be defined as the percentage of signals that hybridize to the correct locus in a metaphase spread and at no other location7. In our study, all eight probes were shown to have an analytical specificity of 100% (with upper and lower 95% confidence levels of 100% and 98.12% respectively) in a total of 1600 loci that were examined across all eight probe sets8. The high level of analytical specificity seen demonstrates that these probes will allow confident probe signal interpretation with no cross-hybridization.
Analytical sensitivity can be described as the percentage of chromosome targets or interphase nuclei with the expected normal signal pattern in a negative cell sample for that abnormality. This is a measure of how well the probe detects the target sequence7. In this study, the FISH probes were hybridized to normal specimens and the number of expected normal signal patterns seen was expressed as a percentage.
A FISH probe exhibiting good hybridization would be expected to have an analytical sensitivity of at least 95%. All eight FISH probes evaluated in this study showed an analytical sensitivity of greater than 98% indicating that these FISH probes provide reliable, consistent results, enabling fast accurate analysis to be performed.
 Table 2: Analytical sensitivity of the eight FISH probes in this study
Table 2: Analytical sensitivity of the eight FISH probes in this study
The clinical studies compared the incidence rates of each of the positive rearrangements indicated by the eight probes being evaluated, with the incidence rates reported in published literature (Table 3). All the FISH probes evaluated demonstrated the expected incidence when compared to reported values in the literature (within 95% confidence limits).
The USA-LPH 026 AML/ETO study results did not reveal any rearrangements of the target chromosome within one population sample and required an additional clinical evaluation. To assess the clinical performance of the probe, ten known samples containing the rearrangement were blinded into a cohort of 90 additional samples known to be negative for the rearrangement. All samples were identified correctly. The USA-LPH 026 AML/ETO probe met the acceptance criteria of this supplementary study.
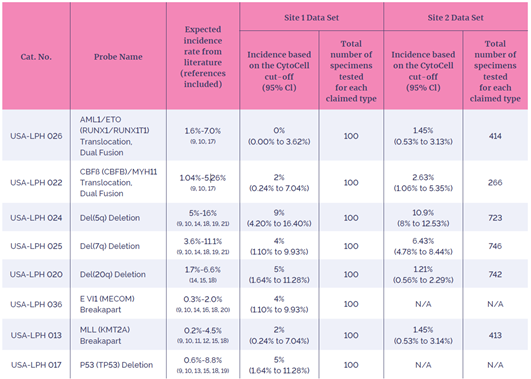 Table 3: Summary of clinical study results. Confirmation that the CytoCell AML/MDS FDA cleared probes detected positive genetic aberrations in concordance with expected genetic aberration prevalence rates from the literature.
Table 3: Summary of clinical study results. Confirmation that the CytoCell AML/MDS FDA cleared probes detected positive genetic aberrations in concordance with expected genetic aberration prevalence rates from the literature.
Reproducibility is a measurement of the consistency of the FISH test when performed at different times by different operators. A highly reproducible test is important to ensure confidence in achieving reliable FISH results with minimal variability in probe performance.
In our study we considered several different types of reproducibility, sample-to-sample (same day), site-to-site, day-to-day and lot-to-lot variability. All eight probes met or exceeded the acceptance criteria of >95% agreement across all variables for intra/inter-day and inter-site reproducibility, demonstrating exceptional consistency (Table 4). Further testing was carried out to demonstrate inter-lot reproducibility.
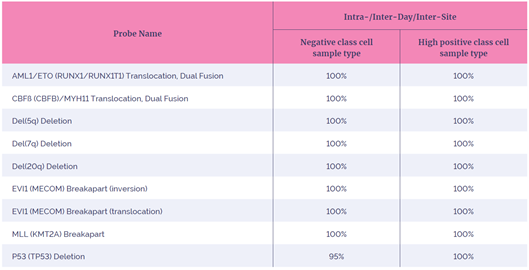 Table 4: Summary of the reproducibility results achieved.
Table 4: Summary of the reproducibility results achieved.
The intra-day reproducibility covered sample-to-sample variability. Inter-day considered variability between separate days. Inter-site reproducibility was measured across three testing sites. Inter-lot measured probe batch-to-batch variability. These variables were jointly analyzed to show an overall reproducibility of these probes. A negative class cell sample is defined as negative for the supported signal pattern. A high positive class cell sample type represents samples that show >45% cell positivity for the supported signal pattern.
The cut-off value (upper reference limit) can be considered as the percentage of scoreable interphase cells with a specific abnormal FISH signal pattern found in a normal cell sample for that abnormality. Good validation practice suggests that cut-offs should be generated from a pool of at least 20 – 25 karyotypically normal bone marrow samples or samples that are negative for the abnormality which the probe is detecting7,22,23,24.
For each probe, varying numbers of samples were used to calculate the normal cut-off values (Table 5). The cutoffsfor all probes were calculated using the BETAINV method, a widely used statistical method for the determination of the normal cut-off 23,24.
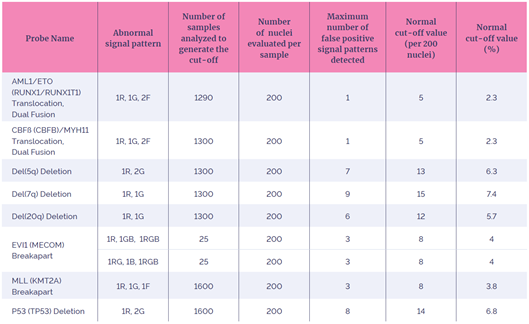 Table 5: Summary of the Normal Cut-Off (or Upper Reference Limit)
Table 5: Summary of the Normal Cut-Off (or Upper Reference Limit)
It is important that any tests used in a diagnostic setting maintain their performance under standard laboratory conditions. Extensive stability studies with a range of temperatures and treatments were performed to assess the FISH probe stability.
It is also sometimes necessary to review slides once the initial analysis has been completed, therefore the assessment of the integrity of final analyzed FISH slides was studied. The stability studies performed are described below and all eight probes maintained their performance.
The FISH probes were stored at -20°C in the dark, an isochronal study was performed where probe lots of different ages were assessed on identical samples. In order to support a 24-month shelf life, the minimum age of lots to be assessed was 25 months; however, lots of probes between 0-34 months were included to provide supplementary data in support of the statistical analysis. All probes met the requirements for a 24-month shelf life claim.
Probes were subjected to 11 rounds of thawing and freezing, subsequent FISH testing showed the probes to be stable after a total of 11 freeze/thaw cycles.
A transport study was conducted at temperatures of 40°C for two weeks, subsequent FISH testing demonstrated stability for all probes.
When stored in darkness at 2-8°C for up to one month, hybridized slides demonstrated reproducible analyzability.
All probes were shown to be stable under limited laboratory light exposure as might be expected during the normal use of FISH probes.
In summary, in order to fulfill the high standards required for an FDA submission, all eight CytoCell FISH probes in this submission underwent stringent performance assessments to underpin the probe performance and ensure that the products were safe and effective for their intended use.
FDA clearance provides you with the confidence that, when choosing FDA-cleared CytoCell probes from OGT, the following benefits are delivered to your laboratory:
Following the extensive analytical and clinical studies, all probes met the stringent acceptance criteria required to demonstrate probe specificity, sensitivity and clinical utility. This provides confidence that these probes can meet their clinical intended use and reduces the validation burden in your laboratory.
The performance of the FDA-cleared AML/MDS probes has been demonstrated to be highly reproducible, delivering high intensity signals with excellent contrast. Consequently, the performance of these probes enables reliable, accurate analysis and low retest rates which can help streamline operations in your laboratory.
CytoCell FDA-cleared probe kits are shipped in an easy-to-use, pre-mixed probe format and are supplied with DAPI, along with detailed protocols and analysis guidelines. The kits help to simplify processing and minimize the chance for error, adding further efficiencies to the FISH workflow.
Our quality products are all backed by expert support from our technical specialists enabling you to focus on delivering high quality, rapid test results to the patient.
For additional details regarding the experimental design and criteria used, please refer to the relevant Package Insert (Instructions For Use, IFU) document provided with the product.
CytoCell® FDA cleared probes: For In Vitro Diagnostic Use. Rx only. Product availability may vary from country to country and is subject to varying regulatory requirements. Please contact your local representative for availability.
