The analysis of structural variants, such as copy number variants (CNVs), is an important aspect of clinical genetics research. Whilst many technologies are used for determining CNVs in human DNA, array comparative genomic hybridisation (aCGH) is now established as the gold standard for detection of CNVs across the entire genome, and is used not just in research but also in clinical applications1.
As microarray technology has evolved, the resolution at which CNVs can be detected has steadily increased. Despite this, the design strategy behind many microarray designs has made it difficult or impossible to find aberrations smaller than ~30kb2, despite the fact that smaller aberrations have been demonstrated to be relevant in Mendelian disorders3. In order to tackle this problem, sophisticated probe design approaches have been developed over the last few years, and have made it possible to increase the resolution of arrays much further – targeting important genetic loci in such a way that CNVs can be found even at the exon-level4.
However, in order to develop arrays that can robustly analyse CNVs at this size, a number of factors need to be taken into account during the design process.
Due to the defined/fixed number of probes on any array, an important aspect of development is to utilise these as effectively as possible. The ability of an array to detect CNVs is defined by the number of probes targeting any given region – these regions might be high-priority regions which need highly clustered probes to deliver desired resolution, or lower priority “backbone” regions which will have less clustered probes. It is therefore important to strike the right balance between probes used across the backbone regions and highly targeted regions to suit the microarray’s purpose. To do this, OGT’s design algorithms work to find the best possible genomic targets for each probe in order to achieve the desired resolution, without compromising on overall content.
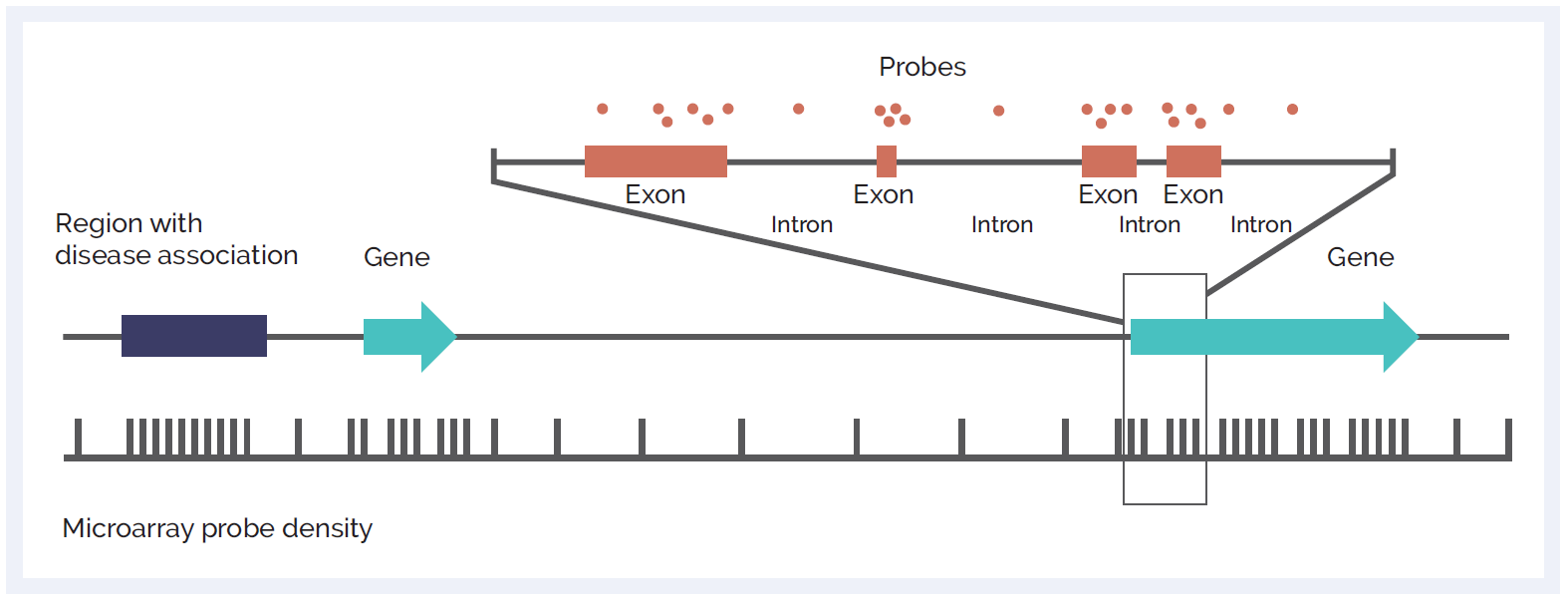
Figure 1: Schematic of microarray probe density across different genetic elements. Probes can be focused across exons to give ultra-high resolution at the exon-level, enabling single exon CNV discovery. High probe density can also be targeted at non-genic regions of importance. Backbone probes, dispersed throughout the genome, allow for discovery of larger variants at standard resolution in other areas.

The key to producing a microarray able to robustly detect CNV at any desired resolution is in the design of the probes themselves and the way they function. To understand what makes a good probe, we need to consider the following:
With all of these factors in mind, a sophisticated approach to probe design needs to be adopted to ensure probes provide the most informative signals and consequently the best possible resolution across the regions of interest. OGT’s design workflow includes all of the following in silico steps to design and identify the best possible probes to target any given region:
In addition to all the in silico steps taken during probe design, all OGT catalogue microarrays also undergo an empirical optimisation process, to ensure that all probes are working at peak performance. At times, selection of the best possible probes from a number of potential options is necessary, especially for regions that are difficult to target due to the nature of the sequence. By testing a number of different probes across a great number of repeats, selection of the best possible probes to target any desired loci is possible (Figure 2). All of this design work leads to better probe specificity, reduced noise, and importantly more accurate results (figure 3).
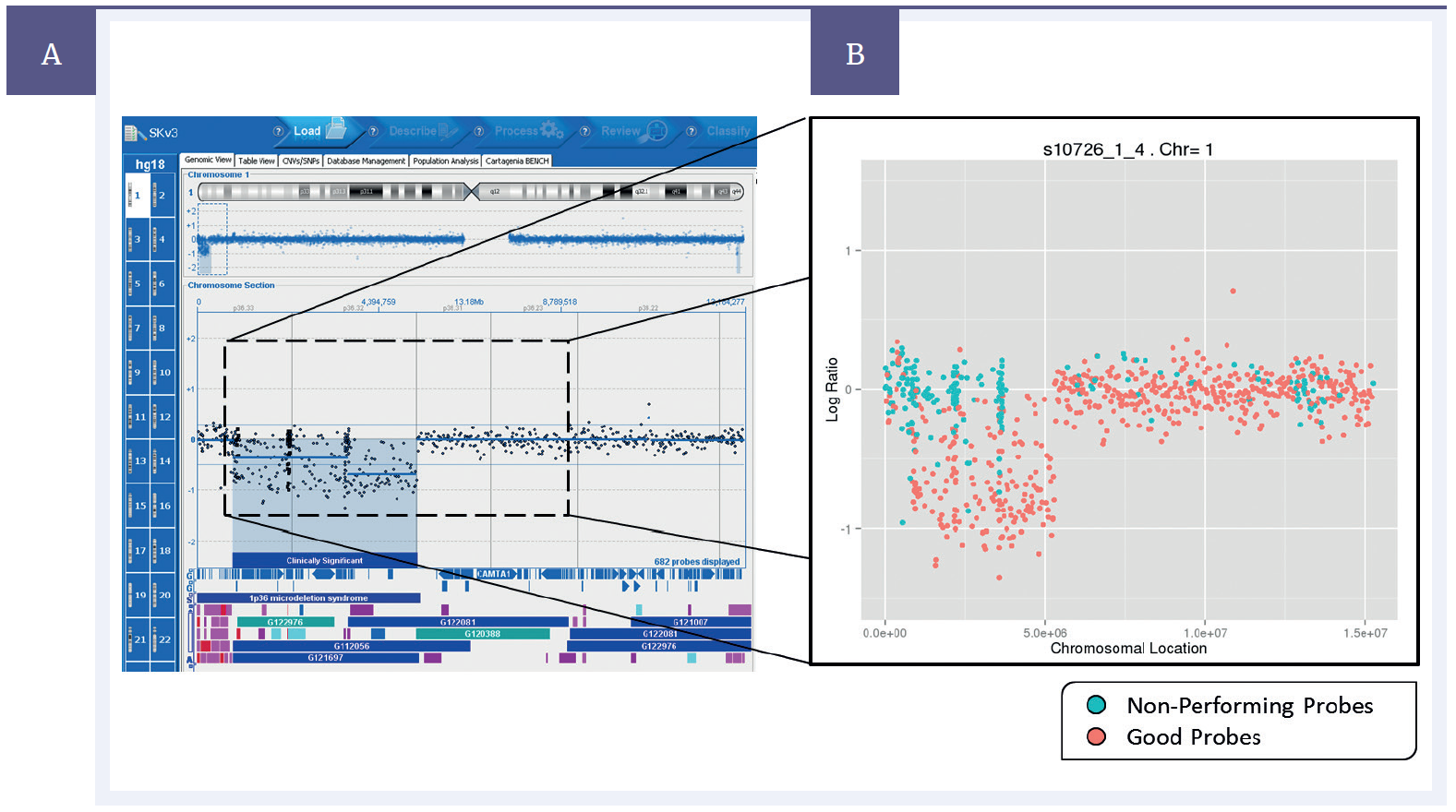
Figure 2: Left – Data from a single experiment with a deletion present on chromosome 1, shown on CytoSure® Interpret Software. Right – average probe performance across 4000 repeats. “Non-performing probes” (blue) defined as those which gave an incorrect result more often than a correct result, “Good probes” (pink) defined as those which gave a correct result more often than an incorrect result.

To aid with the design process, OGT has developed a database of probes called the Oligome™, which includes probes across the entire genome that are in silico or empirically optimised, allowing for the rapid development of microarray designs. Many hundreds of design projects have helped to continuously improve the database, (which now includes over 20 million probes) with the data from these projects improving the design process and algorithms in an iterative fashion.
A number of different factors must be taken into account when designing arrays, especially those that offer exon resolution. OGT is able to offer high-resolution arrays due to the design process developed as a result of many years of experience designing bespoke microarrays.
Moreover, the entire CytoSure workflow is optimised for robust analysis of CNV – to complement the excellent performance of the microarrays, CytoSure Labelling kits offer very low DLRS (derivative log ratio spread – a measure of noise in array data); and CytoSure Interpret software offers a robust, feature rich and user-friendly platform for the analysis of microarray data.
CytoSure®: For Research Use Only; Not for Diagnostic Procedures.
Call +44 (0)1865 856800 Email contact@ogt.com
Send us a message and we will get back to you