Jacqueline Chan, Lyudmila Georgieva, Sabine Eckert, Faidra Partheniou and Graham Speight (OGT, Oxford, UK)
Using OGT’s extensive background in bait design we have developed a range of fully tested and optimised baits targeting all coding exons of a range of key cancer-related genes (Table 1).
To evaluate the application of a hybridisation-based approach we:
The SureSeq hybridisation-based approached was used throughout this study; the workflow of this is outlined in Figure 1.
Use of target-capture allows the removal of PCR duplicates which can obscure the minor alleles present within a sample.
To confidently call low level variants, NGS reads need to be evenly distributed across all regions of interest. Uniformity of coverage is a useful value with which to compare this distribution and can be expressed as the percentage of target bases that have greater than 20% of the mean coverage.
As reported extensively in the literature1-3, the uniformity of coverage from captured-based approaches such as SureSeq consistently outperforms those enriched using an amplicon method (Figure 2). Furthermore, in our sample set we found the high levels of uniformity are maintained when starting with ~250 ng DNA (light blue bars).
The uniformity of coverage for most samples is greater than 99% of bases covered at 20% of the mean, ensuring that all bases within the panel can be confidently assessed.

Figure 2: Assessment of the uniformity of coverage from FFPE-derived DNA using an amplicon and the SureSeq hybridisation capture-based approaches. Enrichment by SureSeq sequence capture (dark blue bars) demonstrates better uniformity than that of an amplicon-based approach (white bars). The level of uniformity is maintained when starting with ~250 ng DNA (light blue bars). Samples are ordered by increasing DNA Integrity Number (DIN) determined by Agilent 2200 TapeStation – value in brackets.

The superior uniformity of coverage of key exons in BRCA1, and BRCA2 from Sample 6 with SureSeq compared to an amplicon-based panel is shown in Figure 3. Regions of low complexity such as repetitive sequences and high/low GC content often appear skewed in NGS data. However, as a hybridisation capture-based approach will also provide data from the flanking regions adjacent to the target the coverage profile is “smoother” than seen in panel iii. This enables reliable identification of somatic single nucleotide variants (SNVs) and indels in solid tumour samples.
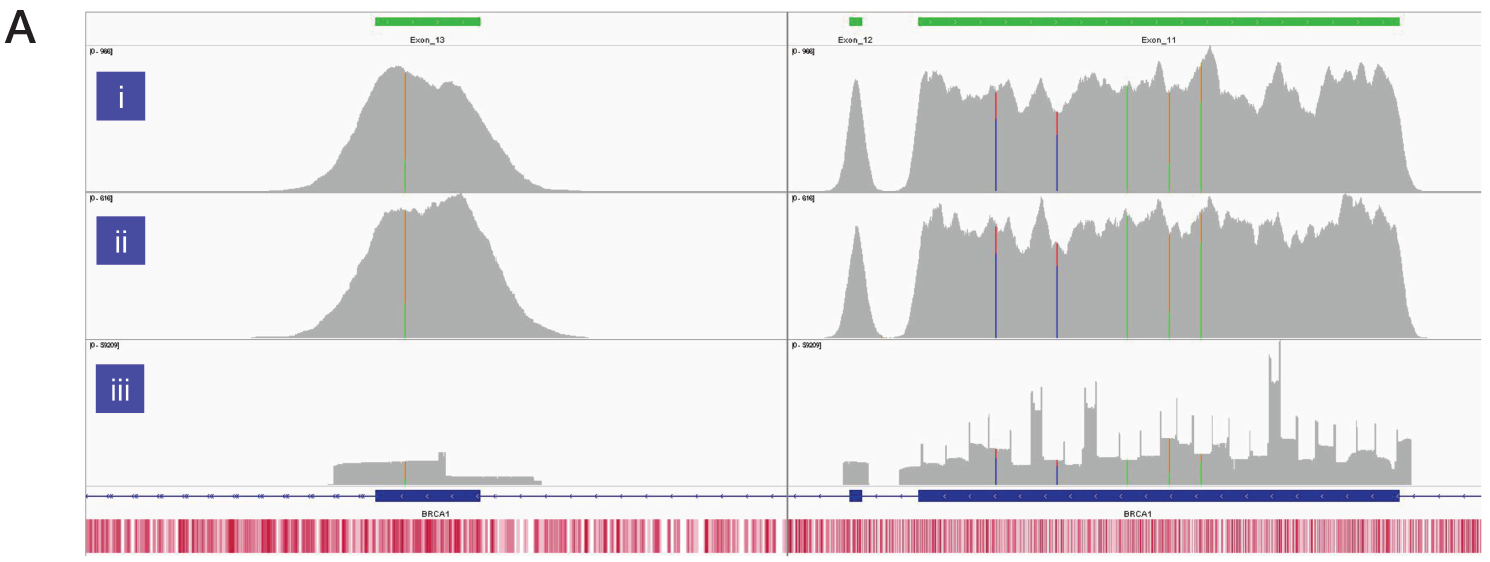
Figure 3A: Assessment of the uniformity of sequencing coverage from FFPE-derived DNA using an amplicon-based and the SureSeq hybridisation-based capture approaches. Integrated Genomics Viewer4 images comparing of depth of coverage of BRCA1 exons 11-13. Panels i and ii - SureSeq Ovarian panel starting with 600 and 300 ng of FFPE-derived DNA respectively. Panel iii – data from an amplicon-based approach. Areas circled in red highlight target regions of very low coverage in the amplicon-based panel. Depth of coverage per base – grey; targeted region – green; gene coding region as defined by RefSeq – blue; GC percentage – red.

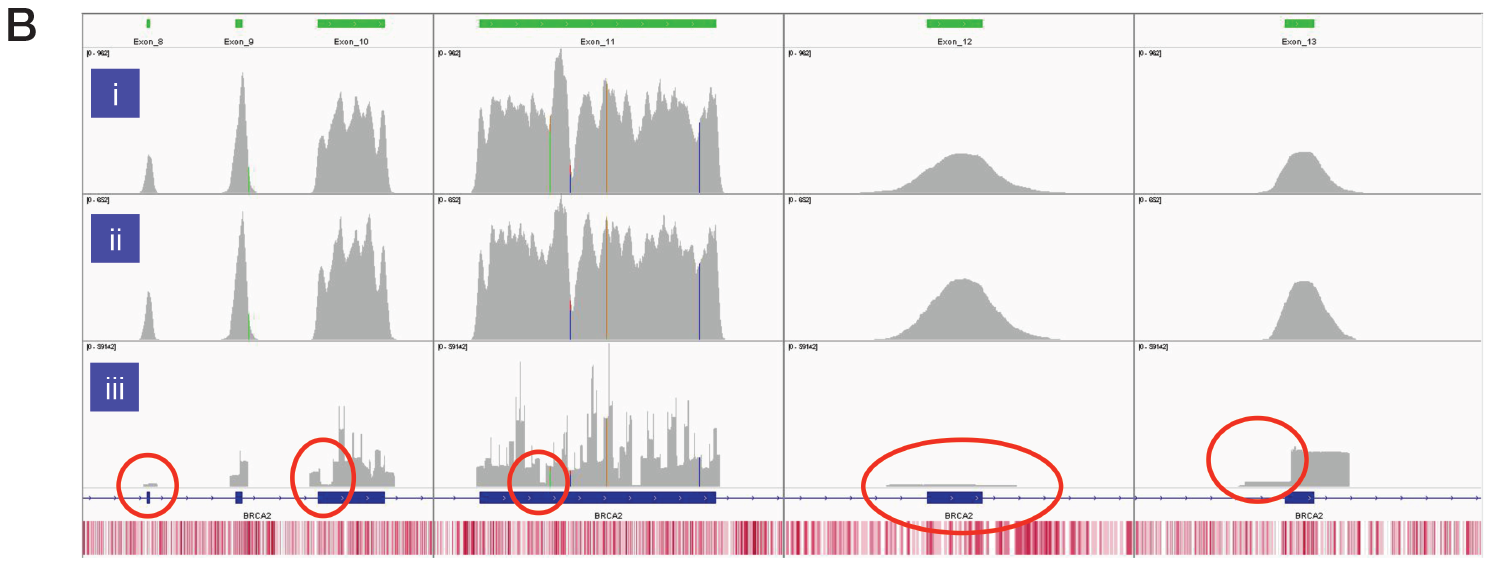
Figure 3B: Assessment of the uniformity of sequencing coverage from FFPE-derived DNA using an amplicon-based and the SureSeq hybridisation-based capture approaches. Integrated Genomics Viewer4 images comparing of depth of coverage of BRCA2 exons 8-13. Panels i and ii - SureSeq Ovarian panel starting with 600 and 300 ng of FFPE-derived DNA respectively. Panel iii – data from an amplicon-based approach. Areas circled in red highlight target regions of very low coverage in the amplicon-based panel. Depth of coverage per base – grey; targeted region – green; gene coding region as defined by RefSeq – blue; GC percentage – red.

The high mean target coverage and uniformity of coverage was maintained across the two sample types and different starting amounts of gDNA (Table 2) which ranged from 1000 to 45 ng of DNA.
All samples had 100% concordance for reported SNVs and all allele frequencies were within 5% of the expected value (Figure 4).
SureSeq: For Research Use Only; Not for Diagnostic Procedures.
* Samples kindly provided by Prof. Charlie Gourley (Cancer Research UK Edinburgh Centre)
Call +44 (0)1865 856800 Email contact@ogt.com
Send us a message and we will get back to you