Nikaoly Ciriaco1, Esther Zamora3, Santiago Escrivá3, Rosa Somoza1, Javier Hernández-Losa1, Santiago Ramón y Cajal1, Martín Espinosa-Bravo2, Kaja Wieghardt4, Hillary Sloane4, Anna Starus4, Frank Holtrup4, Johannes Fredebohm4, Daniel L. Edelstein4, Lucy Georgieva5, Graham Speight5, Frederick S. Jones4, and Vicente Peg1
1Pathology Department, Hospital Universitario Vall d’Hebron, Barcelona, Spain; 2Breast Surgical Oncology Department, Hospital Universitario Vall d’Hebron, Barcelona, Spain; 3Oncology Department, Hospital Universitario Vall d’Hebron, Barcelona, Spain, 4Sysmex Inostics, Baltimore , USA, 5Oxford Gene Technology, Oxford, UK
Neoadjuvant treatment (NAT) is being used widely to eliminate tumor burden in breast cancer (BC) patients; the primary goal of such treatments is to reduce the cancer prior to surgery. Although the standard of care is to perform surgery of primary BC after NAT, it is well known that for certain patients achieving clinical complete response (cCR) and pathological complete response (pCR), surgery following such treatment may be unnecessary.
Previous studies have shown that levels of circulating tumor DNA (ctDNA) during therapy and post-surgery can stratify patients that exhibit effective responses vs. those showing minimal residual disease. In this study, we performed longitudinal tracking of plasma TP53 and PIK3CA mutations pre-specified from NGS analysis of tumor tissue specimens from HER2-positive (HER2) and triple negative (TN) BC patients. The primary objective of this study was to assess ctDNA clearance during NAT as a correlate to effective response to treatment, as benchmarked by clinical complete response (cCR) and pathological complete response (pCR). To accomplish this, a prospective study was conducted to identify patient-specific PIK3CA and TP53 mutations in tissue using SureSeq™ NGS technology, which could then be used to track the presence/absence of mutations prior to, during, and following NAT using Sysmex SafeSEQ technology.
In total, 29 TN and HER2-positive BC patients were examined in this study. Tumor tissue samples from all patients were first examined for mutations in both the PIK3CA or TP53 genes using SureSeq technology (Oxford Gene Technology) after isolation of DNA using Qiasymphony DNA Tissue Kits. Individualized plasma ctDNA detection assays using SafeSEQ technology (Sysmex Inostics) were then applied to detect the specific mutations identified in tissue. Mutation detection was carried out in serial plasma samples in each BC patient at baseline prior to NAT, at treatment mid-point, and at post-treatment immediately prior to surgery. ctDNA analyses were performed only on patient samples in which a specific TP53 or PIK3CA mutation had been initially identified in that patient’s tissue. All patients underwent a surgical excision after NAT. Patient characteristics are shown below in Table 1.
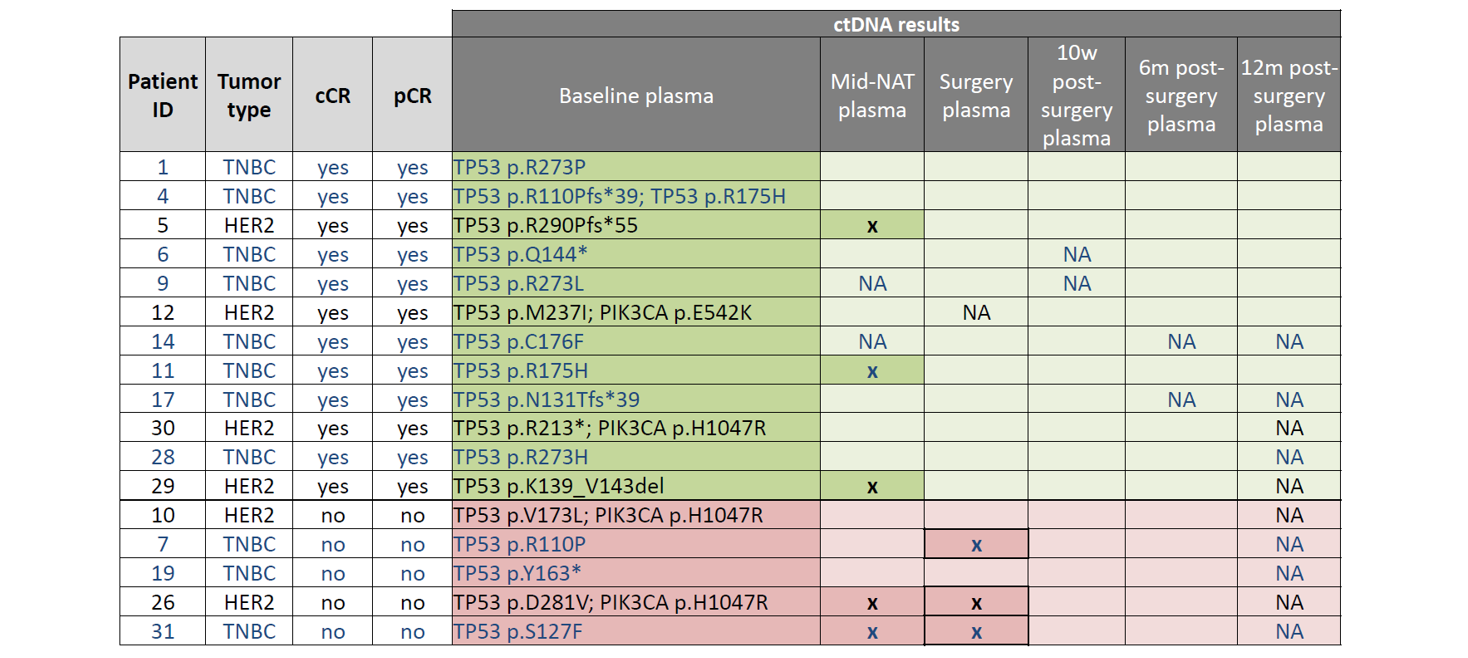
Table 2: Results of longitudinal ctDNA analysis. TP53 and PIK3CA mutations identified in plasma at baseline (pre-NAT) are listed under “baseline plasma” in the table above. These mutations were assessed again at multiple time-points: mid-NAT, post-NAT (“surgery”), and at 3 time-points post-surgery (10w, 6m, and 12m). Samples with ctDNA detected are indicated by an “x”, whereas samples determined to be ctDNA negative are indicated by blank cells. “NA” denotes samples that were not available for analysis. TNBC patients indicated in blue text; HER2+ patients indicated in black text. Green highlighted cells indicate patients with cCR; Pink highlighted cells indicate patients with NO cCR.

SureSeq™: For Research Use Only; Not for Diagnostic Procedures.
Call +44 (0)1865 856800 Email contact@ogt.com
Send us a message and we will get back to you