Fusion genes, hybrid genes formed from two previously independent genes, are implicated in a wide range of cancers — particularly myeloid cancers. Research has revealed the presence of fusion genes in ~41% of acute myeloid leukaemia (AML) and 29% of acute lymphoblastic leukaemia (ALL) cases1,2.
Traditional methods to detect fusion genes include fluorescence in situ hybridisation (FISH) and reverse transcription polymerase chain reaction (RT-PCR). More recently, next-generation sequencing (NGS) has emerged as a key method for fusion gene identification and characterisation. This technique, unlike FISH, can simultaneously detect multiple fusion genes in a single assay. Furthermore, hybridisation-based NGS assays, such as the SureSeq™ Myeloid Fusion Panel, can identify novel fusion partners — providing more comprehensive and informative analyses than possible using PCR-driven methods.

Combining expert-led panel design with hybridisation-based enrichment for unparalleled uniformity and depth of coverage

Detect 30 common myeloid fusions plus novel fusion partners in a single cost-efficient assay

Streamlined library prep and intuitive, complimentary data analysis software — all backed up by OGT’s expert support
The RNA-based SureSeq Myeloid Fusion Panel has been designed in collaboration with leading myeloid cancer experts and has been informed by the latest WHO guidance to include clinically relevant fusions in AML3. The panel further enables the identification of gene expression changes in MECOM, supporting the identification of GATA2-MECOM (inv(3)(q21.3q26.2) and RPN1-MECOM (inv(3)(q21q26).
Figure 1 shows the consistent and confident detection of MECOM overexpression in [A] serial dilutions of HNT-34 cell line as well as [B] research and commercial samples, including positive and negative controls. MECOM expression is normalised to the expression of housekeeping genes and expression values are calculated as counts per million (CPM). ‘Research (+)’ refers to research sample containing GATA2-MECOM (inv(3)(q21.3q26.2)). ‘Commercial (+)’ refers to Universal Human Reference RNA (UHRR) used as positive control. ‘Research (-)’ refers to blood extracted RNA with no GATA2-MECOM (inv(3) (q21.3q26.2)). ‘Commercial (-)’ refers to normal human lymphocyte RNA used as negative control. Error bars represent standard deviation.
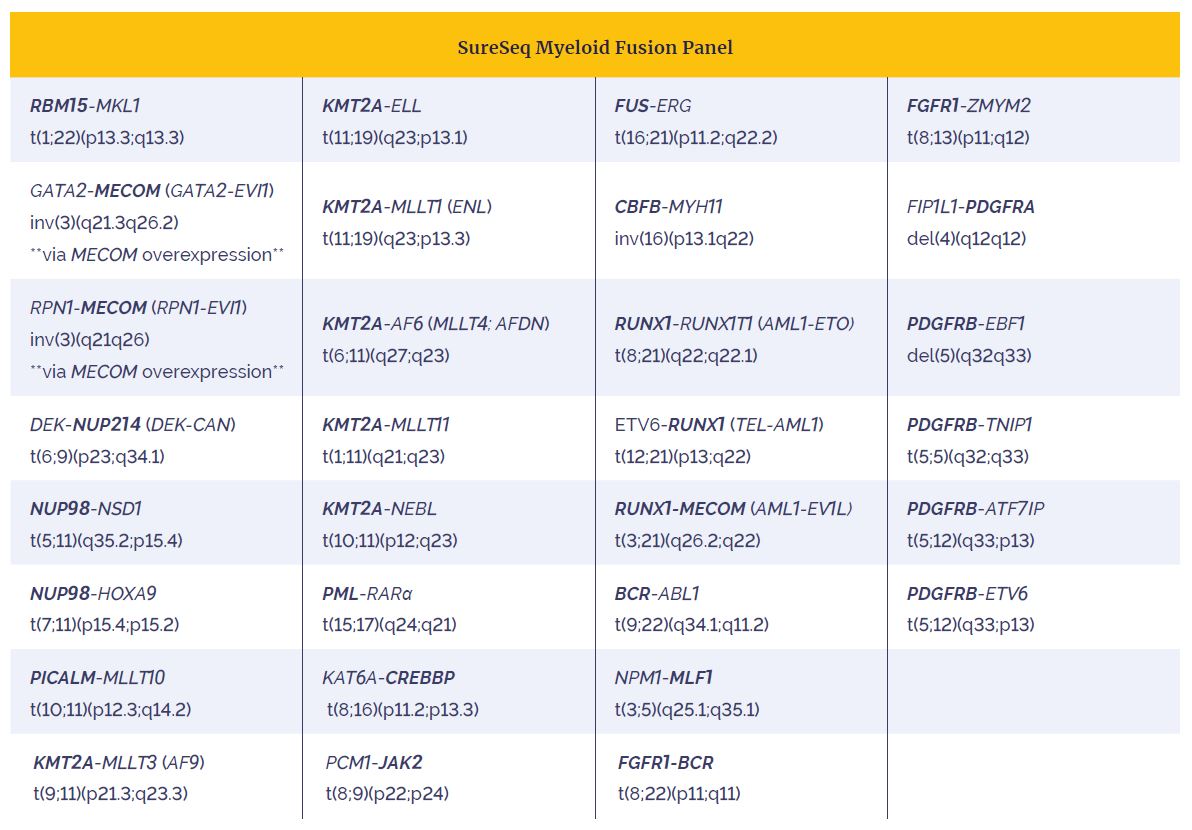
Table 2: The SureSeq Myeloid Fusion Panel allows identification of 30 of the most clinically-relevant fusions implicated in AML. Specific gene targeting (bold text) allows accurate partner gene agnostic identification of fusion genes.

By harnessing RNA-based partner gene agnostic technology you can simultaneously interrogate baited target fusions, including driver genes with multiple fusion partners (i.e. KMT2A).
Together, with the ability to identify novel and/or rare fusions, this panel fully supports your research into myeloid cancer classification and progression. This unique approach further serves to minimise the panel size, lowering sequencing costs and enabling increased depth of coverage for more sensitive results.
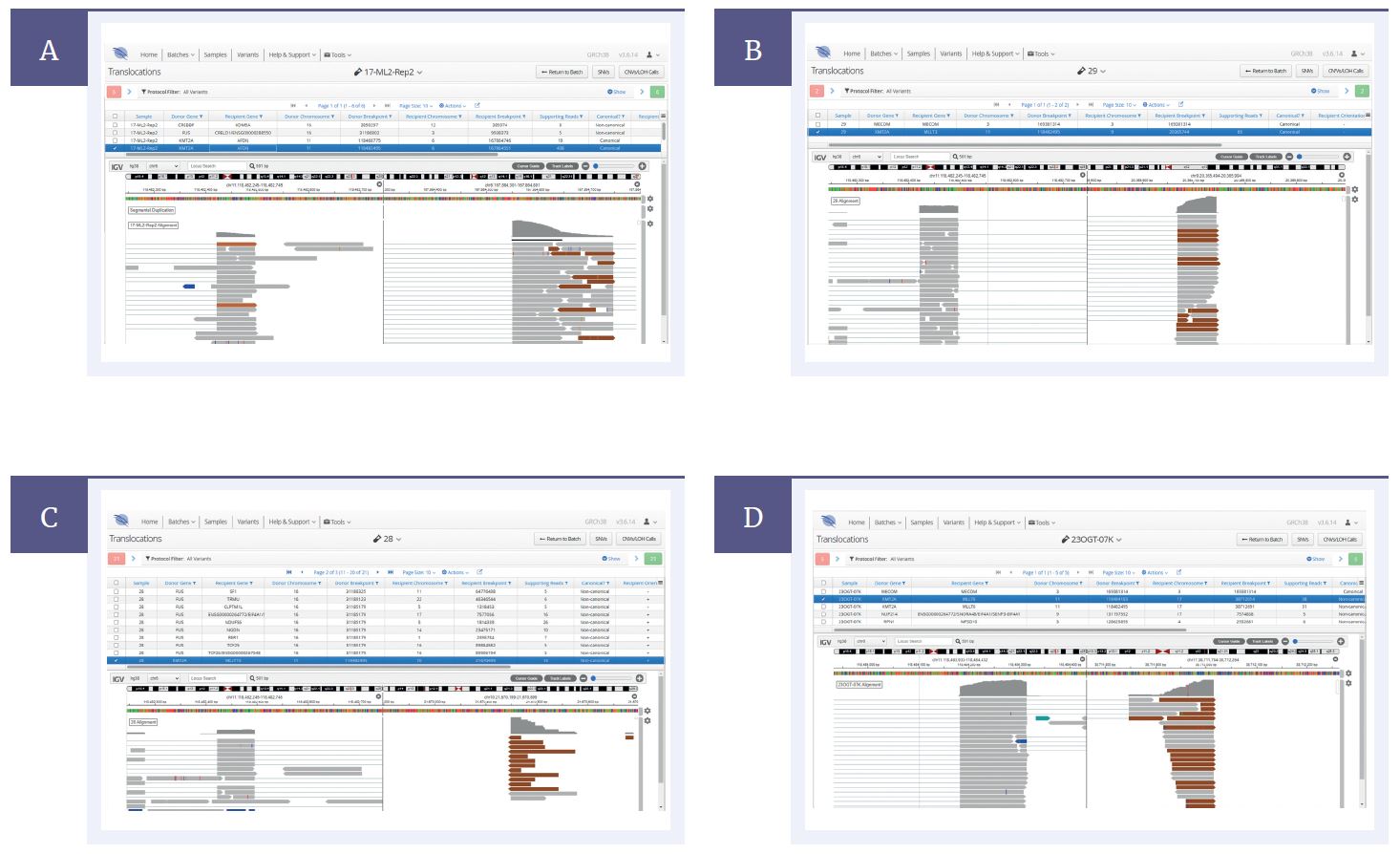
Figure 2: Interpret, OGT’s complementary analysis software, enables identification of ( [A], [B] ) canonical fusions such as KMT2A-AFDN (MLLT4) and KMT2A-MLLT3 as well as ( [C], [D] ) novel fusions such as KMT2A-MLLT10 and KMT2A-MLLT6 emphasising the partner gene agnostic capability for fusion detection.

The SureSeq Myeloid Fusion Complete NGS Workflow Solution V2 is an RNA-based assay, which allows sensitive and more cost-efficient identification of fusion genes than alternative DNA-based panel designs. Only transcriptionally expressed and therefore more clinically relevant gene fusions are analysed.
All SureSeq panels utilise hybridisation-based enrichment, which, when combined with OGT’s intelligent probe design, offers highly uniform coverage for sensitive and reliable results.
Two kit sizes are available, offering the facility to analyse either 24 or 96 samples, with multiplexing of up to 24 samples in a single MiSeq® sequencing run. The streamlined workflow, which utilises the industry-standard Universal NGS Workflow Solution, incorporates Unique Dual Indexes (UDI) prior to sample amplification to support accurate sample demultiplexing for highly robust and reliable results (Figure 3).
Included as standard with all SureSeq panels, Interpret NGS Analysis Software, OGT’s powerful and easy-to-use NGS data analysis solution, delivers comprehensive identification of fusion genes, including novel fusions (Figure 4).
The software allows you to easily visualise fusion genes detected, focus in on breakpoints, the number of reads spanning each breakpoint and alignment of sequence reads at nucleotide resolution — for complete confidence in results. In addition, the software reports normalised gene expression levels related to MECOM rearrangements, plus all data files are available for further downstream analysis.
Browse our full range of myeloid panels, including the focused three-gene SureSeq Core MPN Panel and the SureSeq Pan-Myeloid Panel, incorporating key variants in 70 genes implicated in a wide range of myeloid disorders. In addition, the SureSeq Myeloid MRD Panel enables detection of of low-frequency variants down to 0.05% VAF with confidence, even in challenging biomarkers.