OGT’s range of CytoSure® Cancer +SNP arrays combine long-oligo probes for superior copy number variant (CNV) detection alongside single nucleotide polymorphism (SNP) probes – which function using OGT proprietary technology – for accurate identification of loss-of-heterozygosity (LOH).

Allowing the use of any reference sample with no restriction digest

Through design optimisation

Using CytoSure Interpret® software

Across a choice of formats
Array comparative genomic hybridisation (aCGH) using 60-mer oligonucleotide probes has been shown to offer higher signal-to-noise ratios, increased sensitivity and increased specificity compared to other technologies1.
With other platforms, the use of 60-mer technology for LOH analysis typically requires a restriction digest, which can compromise sample quality, limits the target SNPs to those overlapping restriction sites, and requires a genotyped reference for comparison. However, due to OGT’s unique SNP technology (Figure 1), there is no restriction digest required, the most informative SNPs can be targeted and any reference sample can be used (e.g. normal tissue from the same individual to enable constitutional abnormalities to be filtered out).
Combined with the in silico and empirical optimisation carried out across all OGT catalogue arrays as well as easy customisation to include any additional regions of interest, OGT’s Cancer +SNP arrays deliver flexible and robust analysis of CNV and LOH combined in a single assay.
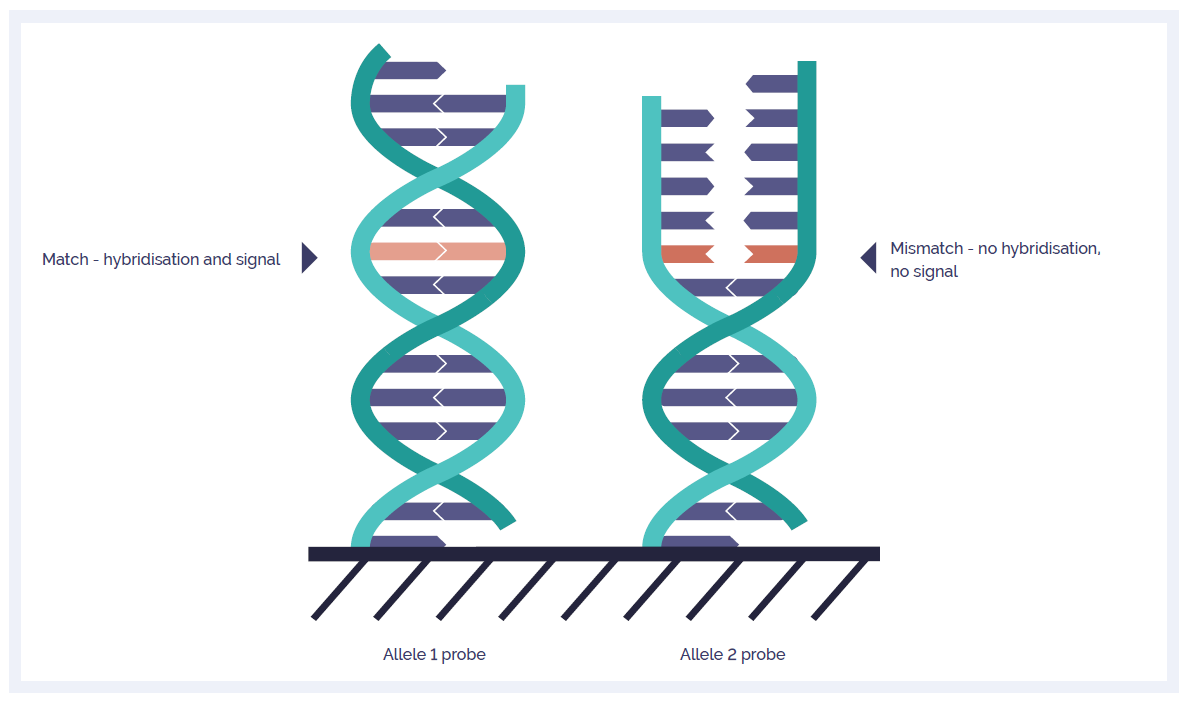
Figure 1. A schematic of the OGT SNP probe technology. Separate probes target both the reference (major) and non-reference (minor) allele. A fluorescent signal is only received from the probe if DNA fragments containing the respective alleles are present in the sample. By comparing fluorescence intensity between the two probes, the allelic status at each SNP position can be ascertained.

The ability of a microarray to detect genetic aberrations accurately is highly dependent on achieving the best possible data quality. OGT leverage years of experience in microarray design to ensure excellent performance in all microarray products. A proprietary pipeline of perl scripts is used to design the best possible probes to target genomic regions of interest, followed by both in silico and empirical optimisation.
Additional targets relevant to your research can also easily be targeted by selecting probes from our Oligome™ database containing more than 26 million pre-optimised probes, including additional SNP probes to improve LOH resolution.
OGT’s CytoSure Interpret Software, which accompanies all CytoSure arrays, is a powerful and easy-to-use package for straightforward analysis of CNV and SNP data (Figure 3), delivering:
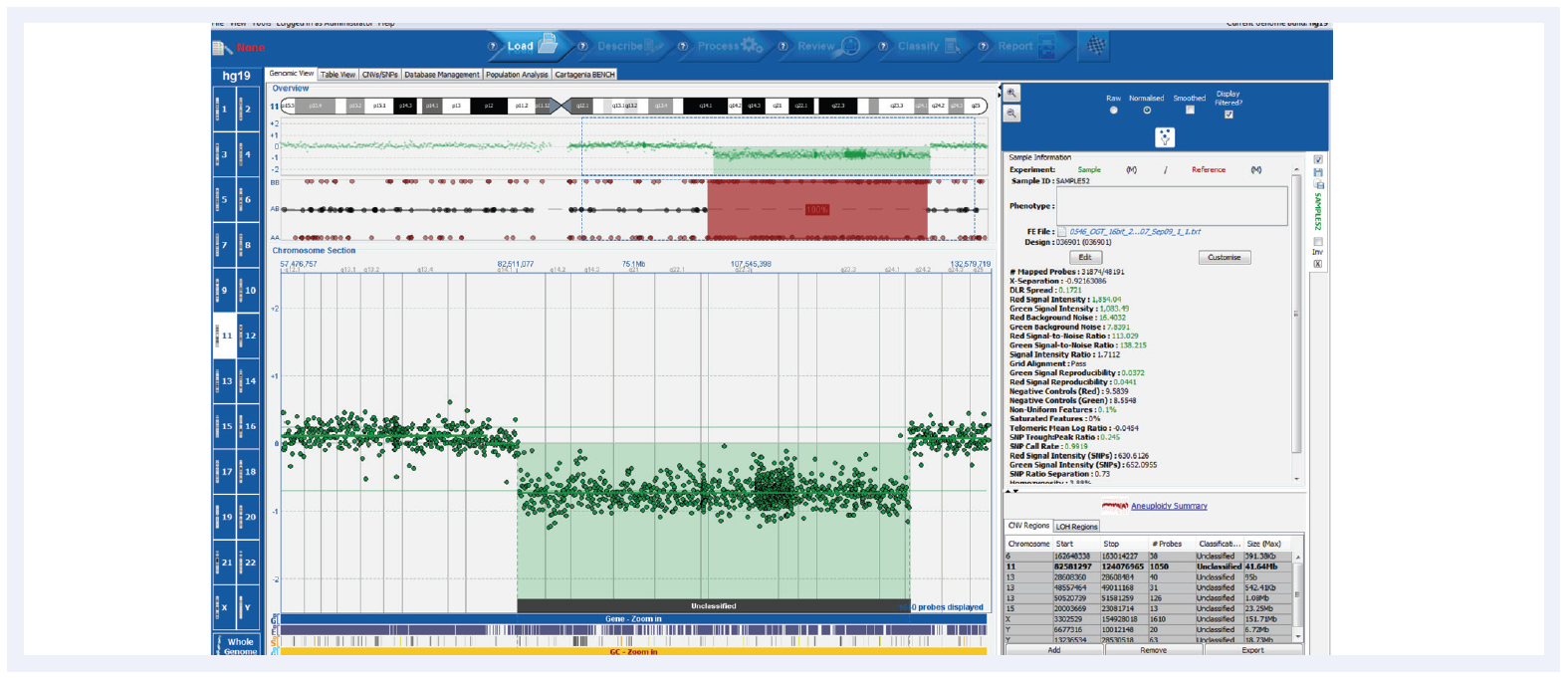
Figure 3. Shown here is a CLL research sample run on the CytoSure Consortium Cancer +SNP array (8x60k) with a deletion and corresponding LOH. CytoSure Interpret offers an intuitive user interface for easy interpretation of genetic findings. Samples kindly provided by Dr Jon Strefford, University of Southampton.

Three fully customisable Cancer +SNP designs are available (Table 1), designed using different formats to suit any analysis and throughput requirement.