Interpret is OGT’s powerful and easy-to-use next-generation sequencing analysis solution, facilitating analysis and visualisation of a wide range of variants and structural aberrations. Coupled with a comprehensive and powerful filtering framework, the software delivers accurate calling of SNVs and indels, as well as structural aberrations, including ITDs, PTDs, CNVs, LOH and translocations. Interpret is designed to work seamlessly with all CytoSure® and SureSeq™ NGS panels and offers flexible accessibility for data analysis; whether through a stand-alone computer1, laboratory server or another web-enabled device. With a wide range of customisation options and links to various mutation databases, Interpret provides effortless translation of all your NGS data into meaningful results.

Reliably call variants ranging from low-frequency SNVs and indels to large structural deletions including CNVs and translocations

Easily customise variant and batch reports and database links to meet the exact needs of your laboratory

Standardise your analysis workflow and overlay bespoke variant filtering to meet your analytical criteria

Log and track user activity and standardise analysis protocols through multiple access permission levels

Optimised for use with SureSeq and CytoSure NGS panels to detect all aberrations covered by your panel
Used in conjunction with CytoSure and SureSeq NGS panels, Interpret facilitates the analysis and visualisation of a wide range of mutation types and structural variants. Complementing the expert panel design and hybridisation-based approach of our NGS panels to deliver unparalleled coverage uniformity, Interpret is integral in facilitating the detection of low-frequency variants consistently and with confidence. Whether your input DNA is high-quality or formalin-compromised, or your research focuses on oncology or rare diseases, Interpret delivers fast and accurate detection of all aberrations covered by your panel (Figures 1-9).
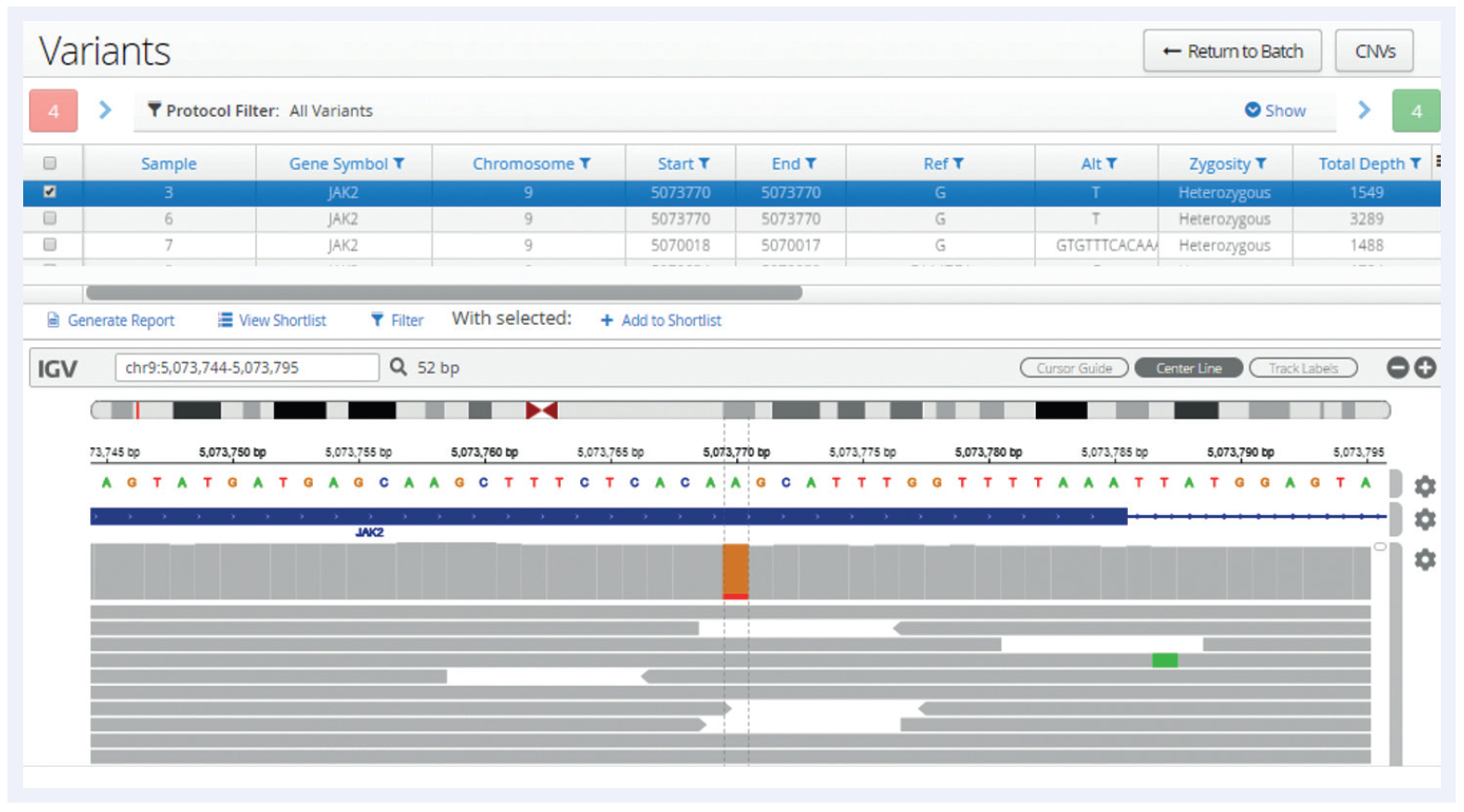
Figure 1: Following analysis, all variants are displayed in a table, below which is an Integrative Genomics Viewer (IGV)2 window allowing a more detailed review of the data and additional verification. In this example a low-frequency JAK2 V617F SNV has been selected and the user is able to view the aligned reads generated by the pipeline.

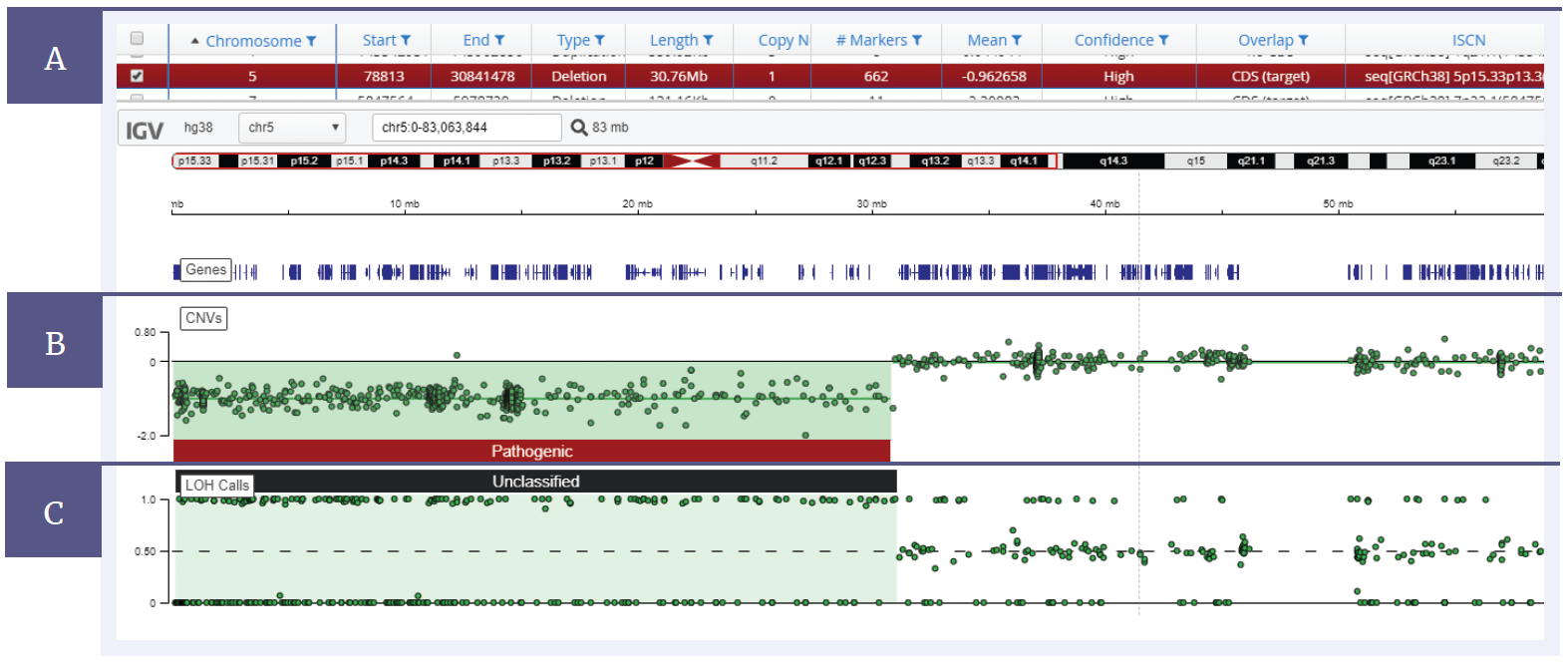
Figure 2: CytoSure Constitutional NGS panel analysed using Interpret, with a 30 Mb deletion on chromosome 5 and LOH across the deletion. [A] Details of the samples and mutations are displayed in a table format. [B] The CNV data is displayed in a log2 ratio plot. [C] A b-allele plot shows the LOH present within the sample.


Figure 3: Detection of FLT3-ITDs of various sizes, including regions containing multiple ITDs. ITD sizes are [A] 174 bp, [B] 225 bp, [C] 195 bp with an additional 6 bp, [D] 120 bp and [E] 168 bp with an additional 69 bp. Note how Interpret can confidently identify even ITDs much longer than the sequencing read length of 150 bp.

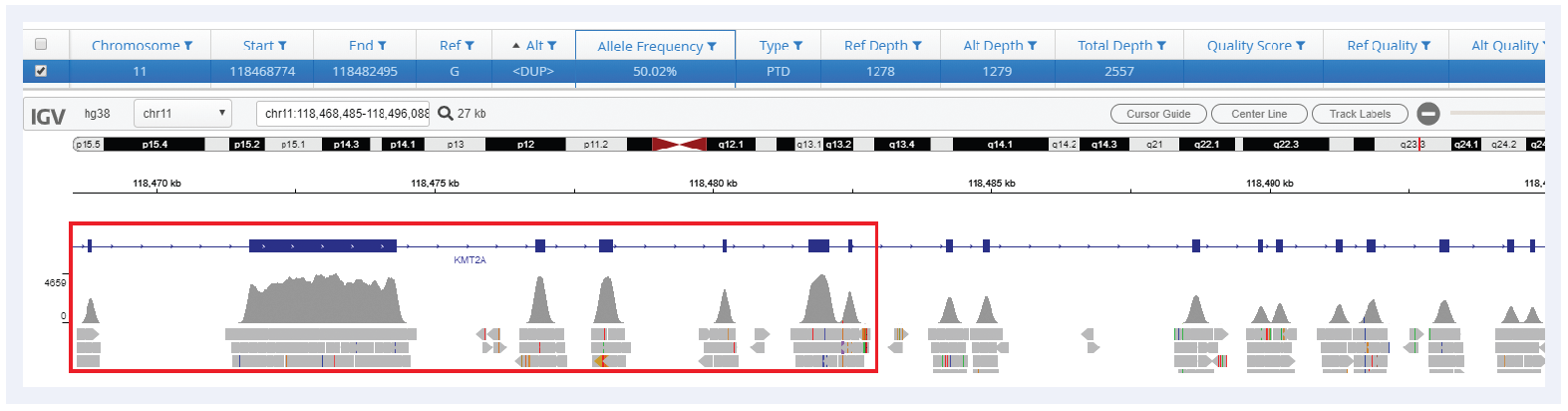
Figure 4: Detection of a KMT2A-PTD spanning exons 2-8. In conjunction with OGT’s expert panel design, Interpret offers robust detection of all sizes of PTDs in KMT2A.

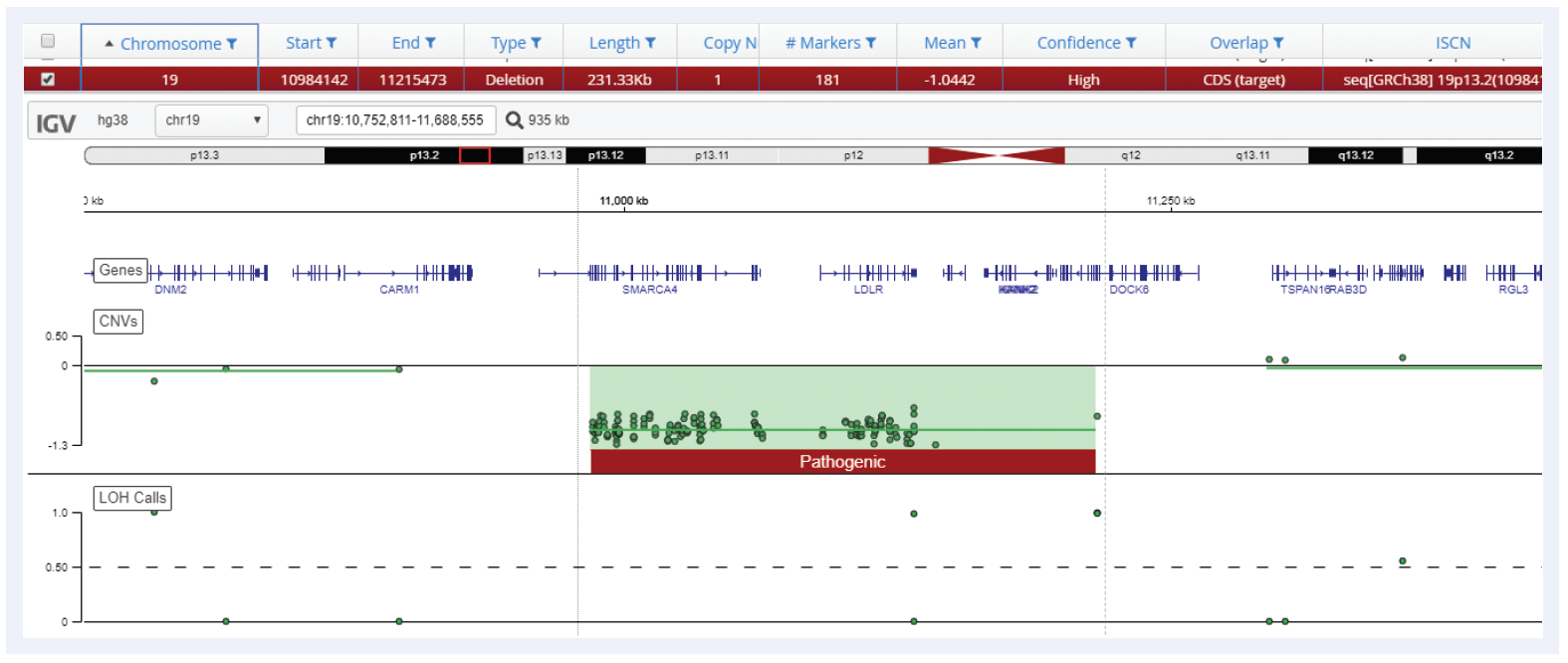
Figure 5: Small 231 kb deletion detected on chromosome 19 using Interpret with CytoSure Constitutional NGS.

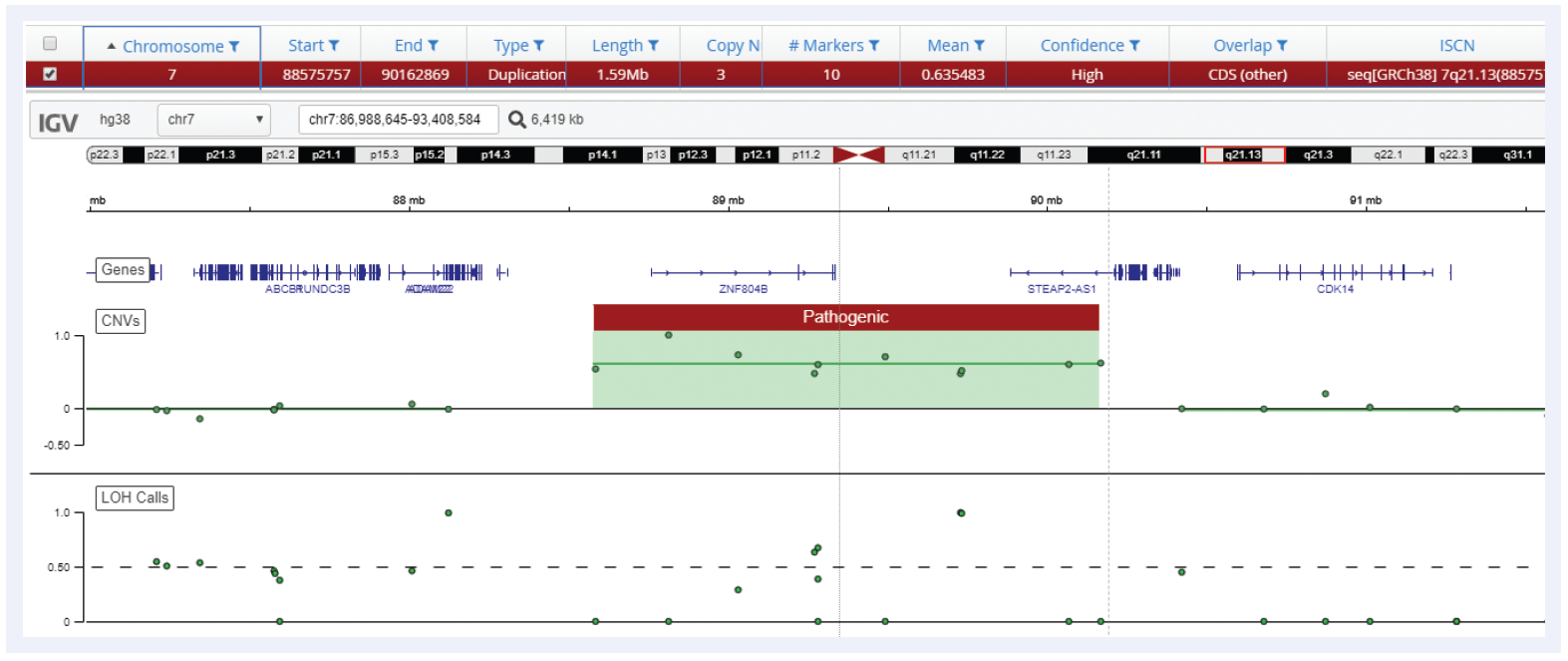
Figure 6: Interpret is able to call duplications with the same precision as microarrays, in this example a 1.59 Mb duplication on chromosome 7 is detected using the CytoSure Constitutional NGS Panel.

OGT has extensive experience in providing individualised options through its established plug-in infrastructure. This enables the software to be tailored to your laboratory’s specific requirements, whether it be report formats, user controls or variant annotation. Additional outputs from the pipeline include a range of publicly available data resources (see figure 10) for annotation of variants detected.
Additional licence-based sources, such as HGMD, can be incorporated on provision of suitable credentials. In Interpret, samples are rapidly processed through customisable protocols and optimised settings in order to generate the variant lists. Your laboratory has the option to use the protocols provided with the software or develop your own through the intuitive user interface. Personal modifications are available for:
Following analysis, results can be viewed in the user-friendly variant browser showing a tabular display of the calls and an IGV window, which can be maximised in a separate window for greater visibility.
Report generation is implemented through a templating system, prepared by OGT following discussion with your laboratory. Our report templates are highly customisable and designed to be modified to individual requirements. For example, one template could simply provide an overview of user activity in the sequencing analysis while another could provide detailed sample or batch analysis results (Figure 11).
An extensive range of dynamic filtering options are available, which allow you to filter your data to meet your exact analytical criteria (Table 1).
Interpret provides a sophisticated user interface which facilitates, through the comprehensive range of filtering options available, the standardisation of your laboratory workflow. These filters can be incorporated into any analysis protocol and are automatically deployed when a particular protocol is selected. This facilitates the building of more complex filter sets to optimise the search and identification of your variants to your exact requirements (Figures 12 and 13).
A relational database stores all activities conducted using the software, from the loading of the samples to the variant calls made in those samples. This facilitates the logging and tracking of individual user activity for consistent data processing and laboratory monitoring.
The database enables implementation of security protocols through multiple access permission levels, allowing users with administrator rights to control all functions of the software and the actions of users based on their roles within the laboratory. Additionally, data stored within the relational database can be easily backed up or ported.

The software provided quick and reliable alignment and variant analysis to interpret results from our core MPN sequencing.
Joshua Landreth
Molecular Laboratory Supervisor, Genetic Associates Inc., USA
The analysis software, provided at no extra cost, offers a user-friendly interface and comprehensive data analysis tools.

Anna Sobczyńska-Konefał
Head of the Department of Hemato-Oncology Diagnostics, Lower Silesian Center for Oncology, Pulmonology and Hematology, Poland