Prostate cancer is now the second leading cause of cancer in men, with recent genome-wide studies helping to clarify the genetic basis of this common but complex disease1. Many of these studies have reinforced the importance of homologous end repair genes including: ATM, BRCA1, BRCA2 and PALB2, in the mechanism of prostate cancer development. Mutations in these genes result in cells having to repair lesions through other non-conservative mutagenic mechanisms.
Choose your ideal prostate cancer NGS panel from our range of fully optimised NGS panel content. Simply mix and match the genes or individual exons you require and get the most out of your sequencing runs. Use in conjunction with the SureSeq™ FFPE DNA Repair Mix† for improved NGS library yields, %OTR (on target rate) and mean target coverage from challenging FFPE derived samples.

Detect low frequency prostate cancer variants consistently with confidence

Get the most comprehensive insight into disease-driving mutations

Easy-to-use analysis solution for accurate detection of all variants
A number of genetic factors have been found that increase prostate cancer risk, including heritable mutations in the genes BRCA1 and BRCA2. BRCA1 is a key player in cellular control systems, having been linked to DNA damage response and repair, transcriptional regulation and chromatin modelling2, while BRCA2 function is linked to DNA recombination and repair processes, being of particular importance in the regulation of RAD51 activity. Figure 1 illustrates the superior uniformity of coverage of key exons of BRCA1 and BRCA2 from an FFPE sample.
PALB2 is a BRCA2 binding protein and the BRCA2-PALB2 interaction is essential for BRCA2-mediated DNA repair. Recently it has been shown that correct PALB2 function is necessary for the homologous recombination repair via interaction with BRCA1, revealing that PALB2 is actually a linker between BRCA1 and BRCA23. The ATM gene, located on chromosome 11q 22–23, includes 66 exons with a 9168 base NGS Custom Prostate Cancer panel 3 pair coding sequence, and encodes a PI3K-related protein kinase (PIKK) that helps maintain genomic integrity. The PI3K-AKT-mTOR oncogenic pathway is frequently enhanced in prostate cancer playing a vital role in development and maintenance4. Figures 2 and 3 illustrate the excellent uniformity of coverage of key exons of PALB2 and ATM, respectively.
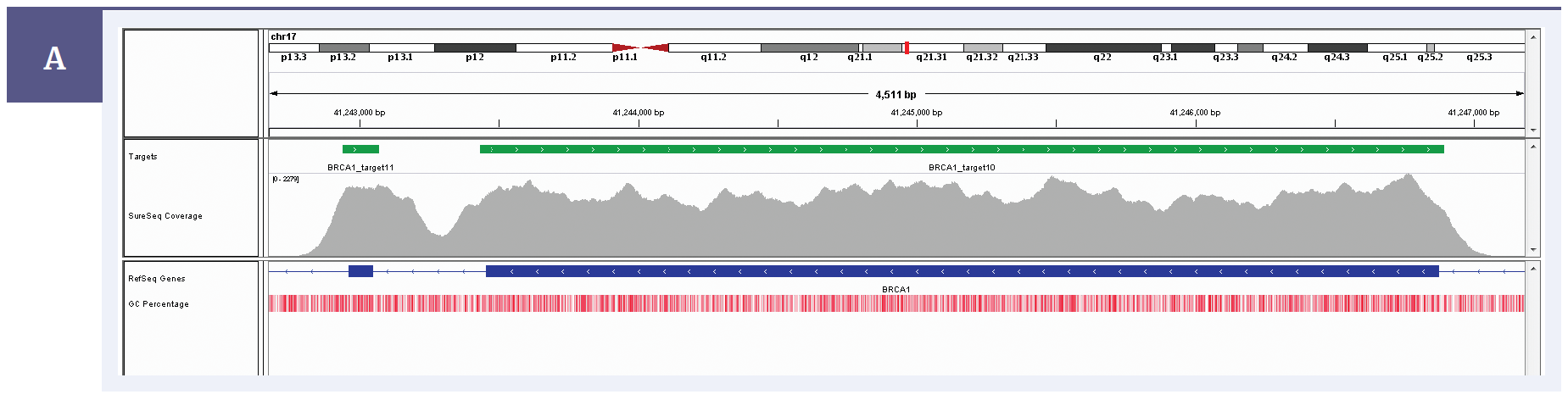
Figure 1a: BRCA1 exon 9 and 10 coverage. Depth of coverage per base (grey). Targeted region (green). Gene coding region as defined by RefSeq (blue). GC percentage (red).

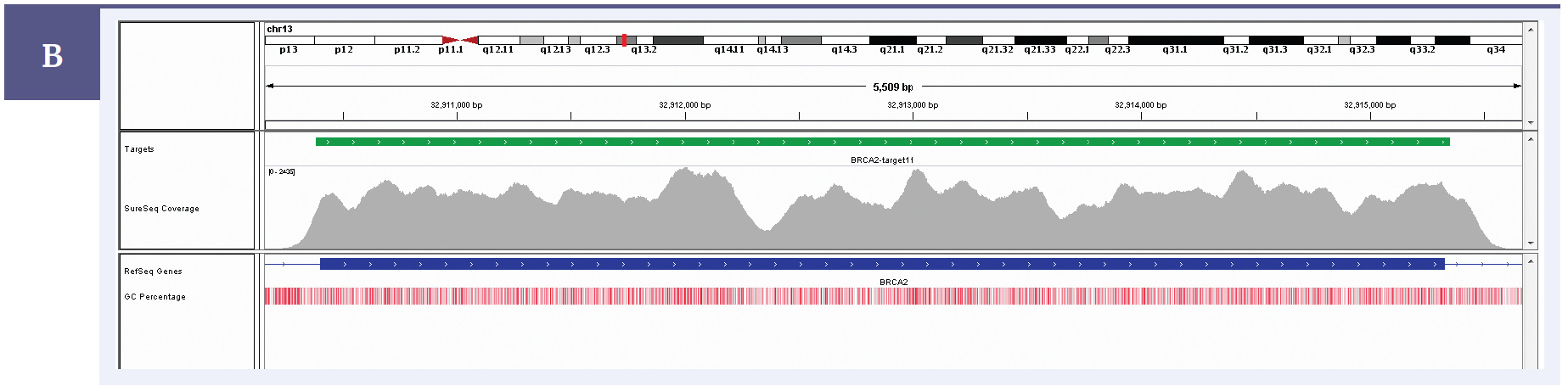
Figure 1b: BRCA2 exon 11 coverage. Depth of coverage per base (grey). Targeted region (green). Gene coding region as defined by RefSeq (blue). GC percentage (red).

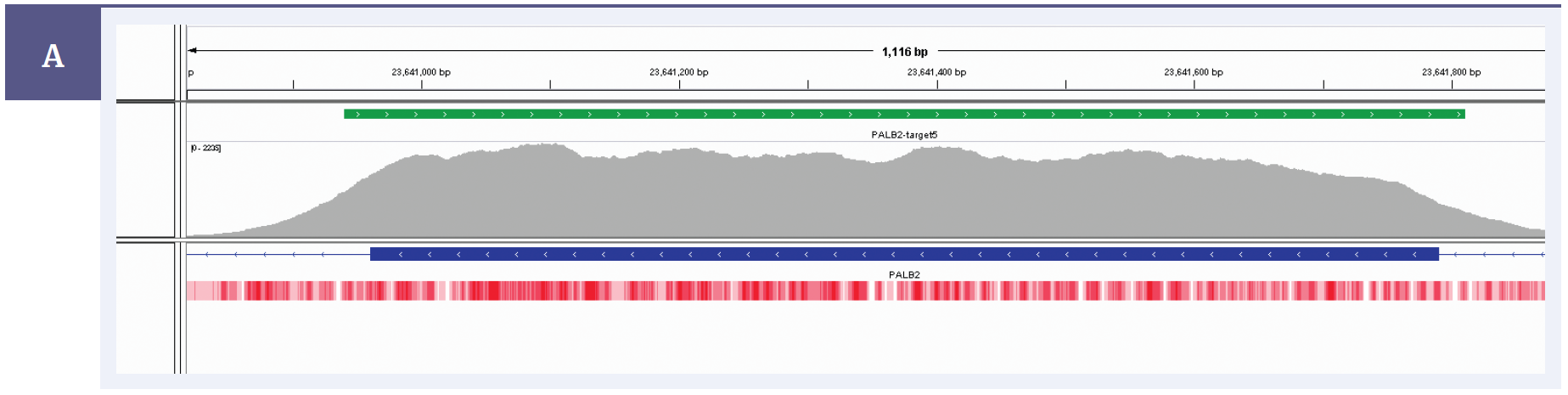
Figure 2a: Illustration of the excellent uniformity of coverage of PALB2 exon 5. Targeted region (green). Gene coding region as defined by RefSeq (blue). GC percentage (red).

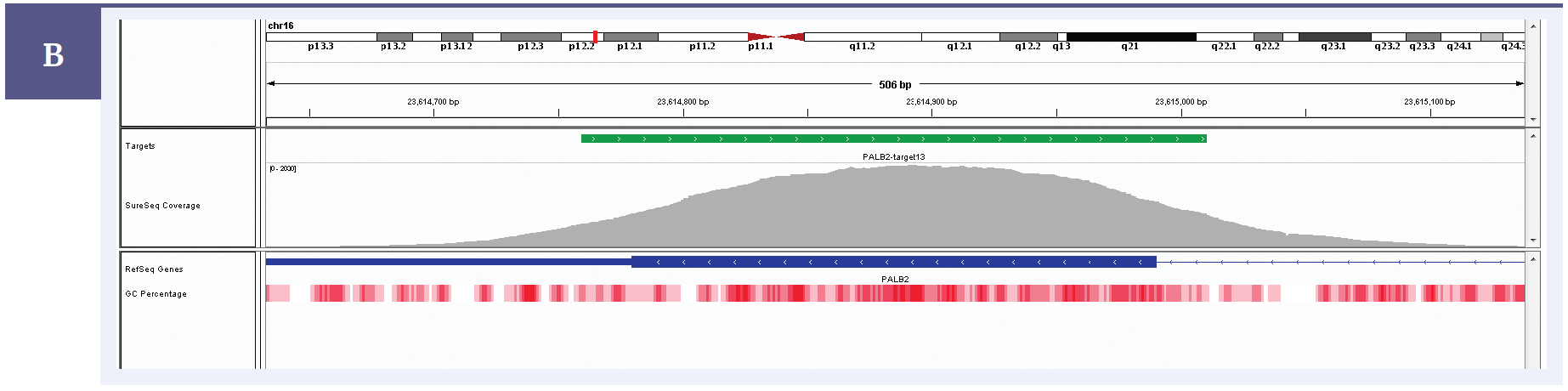
Figure 2b: Illustration of the excellent uniformity of coverage of PALB2 exon 13. Targeted region (green). Gene coding region as defined by RefSeq (blue). GC percentage (red).

* Exon examples not yet available

*Panel size is an estimation based upon recommended custom panel content. †The SureSeq FFPE DNA Repair Mix can only be purchased in conjunction with SureSeq NGS panels, not as a standalone product.