Familial Hypercholesterolaemia (FH) is a genetic condition which results in a high cholesterol level and subsequently leads to a higher risk of early heart disease. It affects approximately 1 in 250 people with around 34 million cases worldwide1.
OGT is offering an optimised NGS panel which has selected the most relevant genes and SNPs implicated in FH, for your research needs. Together with the complimentary Interpret analytical software, the CytoSure® Comprehensive FH NGS platform provides the optimal solution for FH research.

Enabled by the exon resolution of the targeted genes

No more laborious in-house optimisation, decreasing assay development time

Sequence only what’s relevant for your research

Designed to give unparalleled CNV and SNV calling
The hybridisation enrichment methodology, combined with our bait design expertise, allows generation of panels with outstanding completeness and coverage uniformity. Together, this allows the areas of CNV to be easily identified within each sample using our proprietary algorithm — delivering an increased understanding of the sample without an increase in cost or time.
Figure 1 shows CNV in LDLR gene shown using IGV from the Broad Institute.
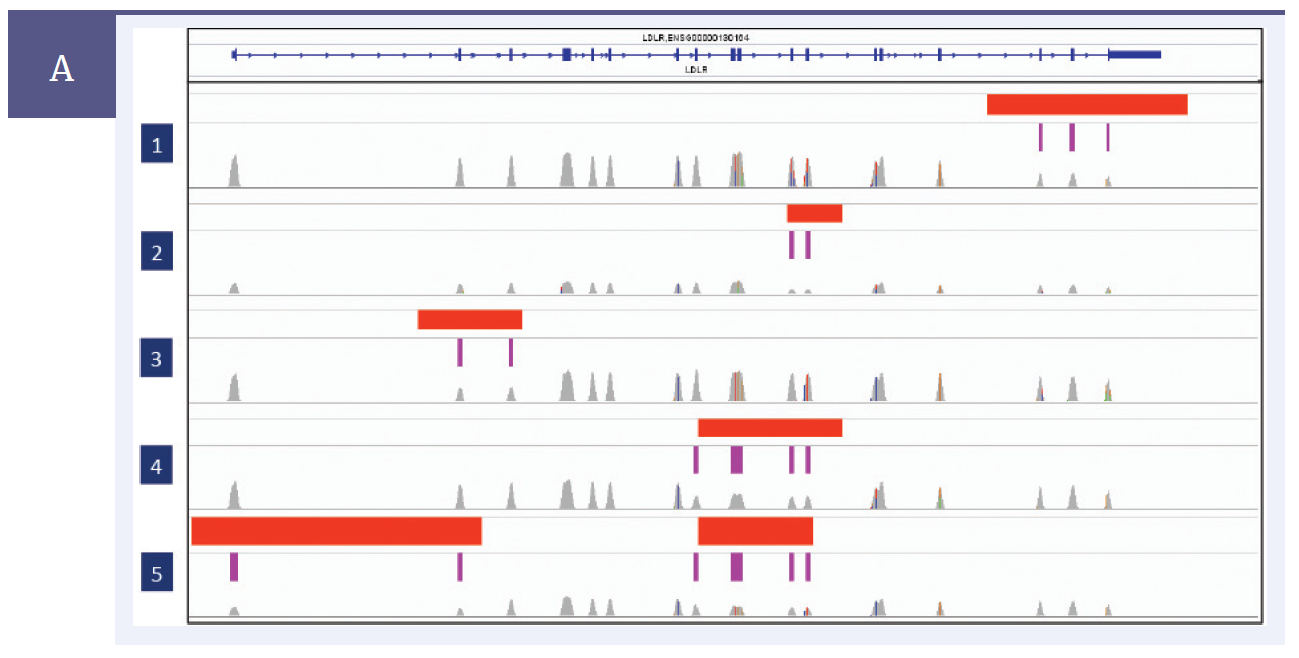
Figure 1a: Red bars indicate areas of CNV (data from aCGH), purple bars represent deleted exons (data from NGS): 5 samples are shown, each with at least one area of CNV. There is complete concordance between the aCGH and NGS data. Note the evenness of the NGS coverage (even peak height) across each exon, allowing the areas of CNV to be easily identified. The data from the custom CytoSure aCGH array, confirms the deletions in LDLR.

The Interpret software has been designed to easily visualise CNVs and SNVs, with an intuitive interface to switch between different sets of results. Interpret also has simple to use protocols and filtering options, to easily target the results of interest.
The CytoSure Comprehensive FH NGS panel has the ability to detect CNVs in whole genes, at exon resolution (Figures 2-3) and can target select SNPs that have been implicated in FH (Figure 4). The CytoSure Comprehensive FH panel can also detect SNVs and Indels within genes, as demonstrated by Figure 5.
* Exon examples not yet available

We were previously using another custom panel but found OGT’s custom FH Panel much easier and more cost-effective to work with. From the start we worked very closely with OGT on the design of the panel and were impressed by the support especially with bioinformatics. The ability to call CNV from the NGS data as well as point mutations is also extremely valuable to us. The fact that the panel is pre-optimised has reduced the need for in-house optimisation and decreased our assay development time.

Dr. Mafalda Bourbon
Head of the Cardiovascular Research Group, National Health Institute Doutor Ricardo Jorge, Lisbon, Portugal