The SureSeq™ NGS Library Preparation Kit generates NGS libraries suitable for the capture of targeted genomic regions using hybridisation. With a streamlined workflow, significantly reduced hands-on steps and hybridisation times as low as 30 minutes, SureSeq offers all of the benefits of hybridisation in as little as a 1-day workflow. For increased convenience and flexibility, the SureSeq workflow can be performed with an enzymatic or Covaris® DNA fragmentation and Speedvac® or bead concentration steps, whilst still delivering libraries of the highest quality. The inclusion of the SureSeq Hyb & Wash buffer, optimised for use with SureSeq NGS panels, simplifies this key step while offering excellent coverage uniformity and reproducibility. Additionally, a new automation workflow enables standardisation of OGT’s library preparation, hybridisation and washing steps for up to 96 samples in a single batch, improving laboratory productivity.

High performance with low duplication rates, high sequence quality and high percentage of on-target bases

All components of the SureSeq Hyb & Wash buffer are ready-to-use with no requirement for multiple wash buffers

Streamlined protocol, minimal manual handling, automation and a rapid hybridisation step offers increased reliability as well as throughput

NGS targeted panel, complete NGS library preparation solution and powerful NGS analysis software are fully optimised for excellent results
NGS data is increasingly being relied upon as a front-line technology for the generation of data for scientific and medical research. A combination of quality metrics are used to give confidence that variants called are real and the number of false positive calls are low. The SureSeq NGS Library Preparation Kit delivers high performance in the quality metrics that really matter, giving more reliable, more trustworthy data.
High duplication rates reduce the complexity of your NGS library and can lead to poor coverage of targeted genomic regions even though average coverage rates appear high.
The SureSeq NGS Library Preparation Kit gives exceptionally low levels of duplication (Figure 1). This ensures more accurate calling, more even coverage and higher levels of confidence in the data produced.
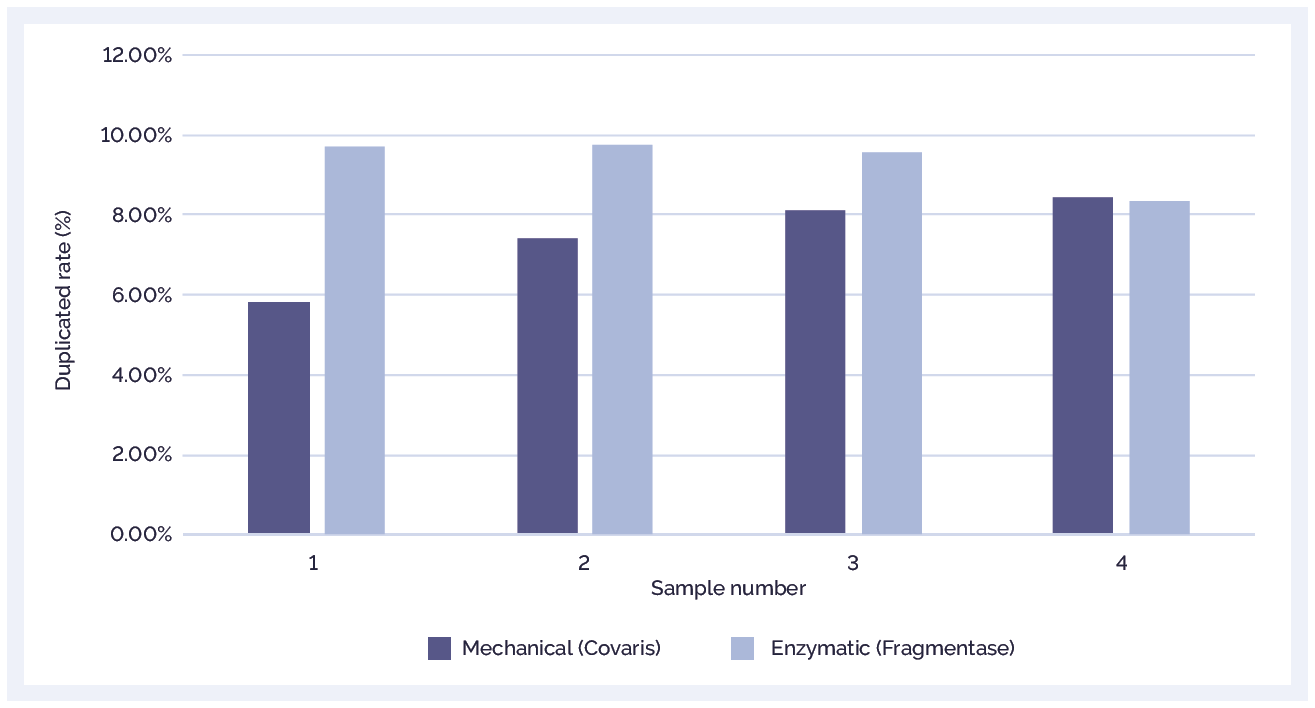
Figure 1: The SureSeq NGS Library Preparation Kit delivers low levels of sequence duplication. The duplication rates are shown for samples fragmented by mechanical or enzymatic methods. Samples were prepared using the SureSeq NGS Library Preparation kit and hybridised with a SureSeq myPanel™ Custom Myeloid 49 Gene Plus panel.

For complete confidence in your data, the sequence quality scores should be high and consistent across the length of each read. The SureSeq NGS Library Preparation Kit delivers exceptional sequence quality scores even 150 bases into the read (Figure 2). This reduces the requirement for post-acquisition bioinformatics, such as read size reduction, to remove poor quality sequence.
In order to maximise the amount of useful information that can be obtained for a given sequencing lane, high coverage depth and low coefficients of variation are desirable. The benefits of superior levels of specific sequence capture include increased sequence complexity and greater depth of coverage of the target regions. The SureSeq NGS Library Preparation Kit delivers excellent reproducibility (Figure 3).
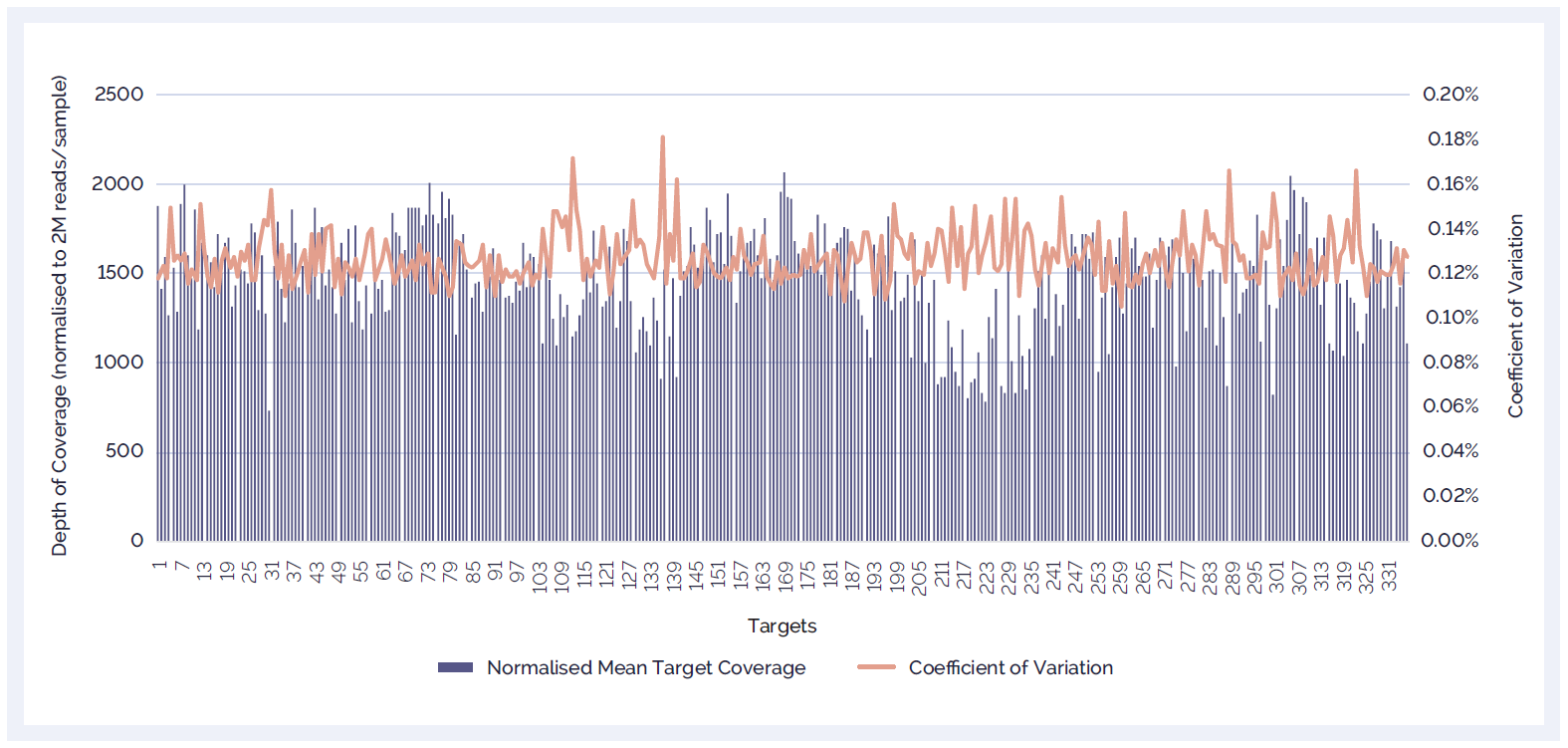
Figure 3: High depth of coverage and a low coefficient of coverage variation from 16 replicates. A SureSeq myPanel Myeloid 49 gene plus and SureSeq Library Preparation Kit were utilised in the evaluation of 336 coding targets in the custom panel (excluded targets on X or Y chromosome) to illustrate the low CV (coefficient of variation) and excellent uniformity of SureSeq.

The SureSeq NGS Hyb & Wash kit is an easy to use, robust and high performing hybridisation solution, containing all the components required to perform the hybridisation and wash steps in SureSeq sequence capture protocols. All components in the kit are ready-to-use, eliminating the requirement to dilute buffers and the possibility of cross-contamination during buffer preparation. There is no need to use multiple wash buffers, as in other manufacturer’s kits, the simplified protocol utilises just a single wash buffer. With hybridisation times as little as 30 minutes, it offers excellent quality sequencing data, when used in conjunction with the SureSeq NGS Library Preparation Kit and SureSeq NGS panels (Figure 4).
By extensive testing of enzymes and buffer optimisation, it has been possible to streamline the standard library preparation protocol to minimise the number of hands-on steps and the overall processing time (Figure 5). For added convenience, OGT now offers Dynabeads™ M270 Streptavidin and AMPure® XP beads for use with SureSeq NGS panels. While the addition of a workflow for automation on the Agilent Bravo A® Automated Liquid Handling Platform offers greater flexibility and productivity for your laboratory.
The SureSeq NGS Library Preparation Kit and Hyb & Wash Kit work hand-in-hand with SureSeq targeted enrichment panels, ensuring you get the most sensitive and reproducible variant detection and industry-leading coverage uniformity (Figure 6).
Interpret is OGT’s powerful and easy-to-use next-generation sequencing analysis solution, facilitating analysis and visualisation of a wide range of variants and structural aberrations. Coupled with a comprehensive and powerful filtering framework, the software delivers accurate calling of SNVs and indels, as well as structural aberrations, including ITDs, PTDs, CNVs, LOH and translocations. Interpret is designed to work seamlessly with all SureSeq NGS panels and offers flexible accessibility for data analysis; whether through a stand-alone computer*, laboratory server or another web-enabled device. With a wide range of customisation options and links to various mutation databases, Interpret provides effortless translation of all your NGS data into meaningful results.
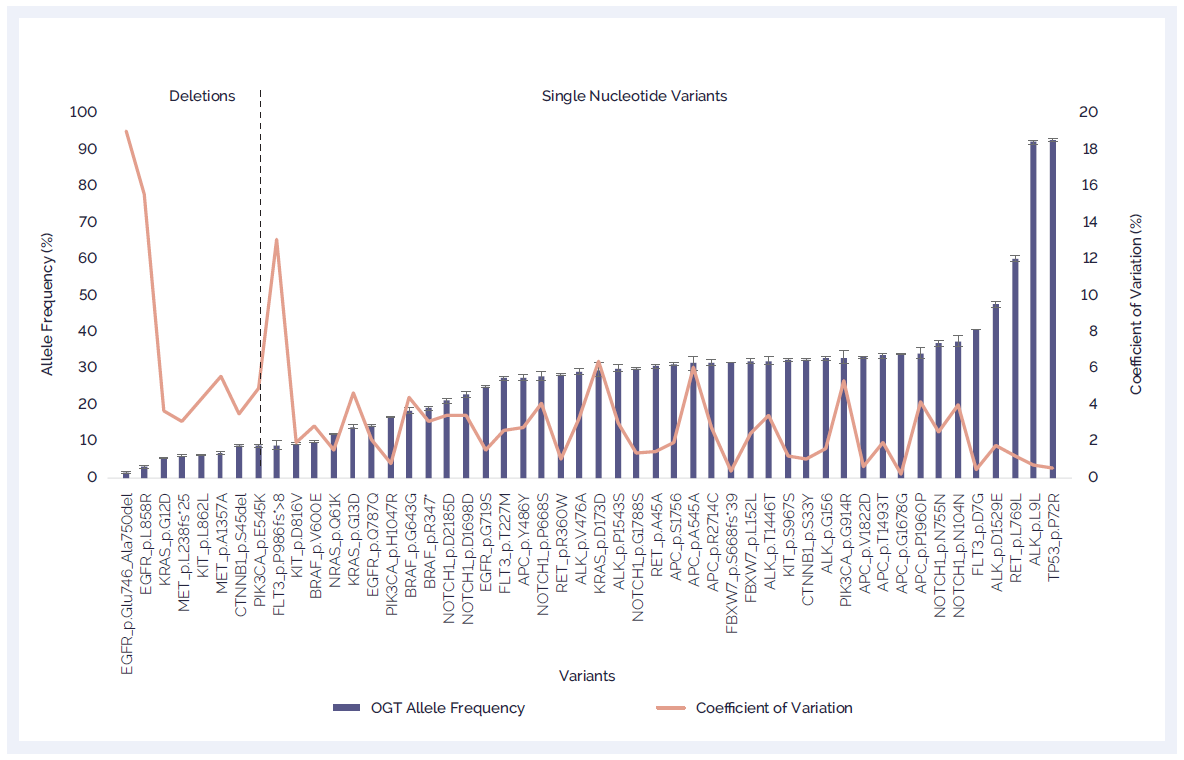
Figure 6. Excellent reproducibility with low allele frequency detection and CV distribution. The SureSeq workflow and software were validated using OncoSpan Reference Standards (Horizon Discovery). A SureSeq myPanel NGS Custom panel was utilised to detect 46 variants in 15 genes present in the OncoSpan Reference Standard. Three different DNA inputs with three independent replicates were processed in parallel. Data generated was analysed using SureSeq Interpret Software.