Connect with our NGS experts today
Want to download this as a PDF? Download now
Contributors
Lyudmila Georgieva, Ezam Uddin, Jacqueline Chan and Graham Speight
Introduction
Myeloproliferative neoplasms (MPNs) are a group of diseases that affect blood cell production in the bone marrow resulting in the overproduction of one or more cell types.
The application of short read NGS for research into MPNs has been held back by the accurate and timely analysis of the key MPN driver mutations. These include: JAK2 V617F and exon 12, MPL W515K/L and S505N, and CALR exon 9 insertion and deletions (up to 52 bp).
Designed for research into the diagnosis, aetiology and prognosis of MPNs, the SureSeq™ Core MPN Panel has been developed by OGT in collaboration with recognized cancer experts to deliver accurate detection (down to 1% variant allele fraction [VAF]) of somatic variants of these key MPN driver mutations.
The aim of this study is to evaluate the SureSeq Core MPN Panel in conjunction with the new streamlined 1-day hybridization-based NGS library preparation kit (LPK).
Methods
Preparation of purified DNA to sequencer-ready libraries in 7 hours and 45 minutes
- An enhanced version of the SureSeq Library Preparation Kit (LPK) was utilized which incorporates an enzymatic DNA fragmentation in combination with a rapid hybridization of just 30 minutes.
- This enhanced protocol reduces the overall processing time by 6 hours, resulting in a streamlined, 1-day workflow that permits the preparation of purified DNA samples to Illumina sequencer ready libraries in a single-day.
- The kit offers a similar turn-around time to amplicon-based enrichment protocols, without the associated disadvantages, such as PCR bias, allelic bias (indels) and drop-outs, as well as poor uniformity of coverage.
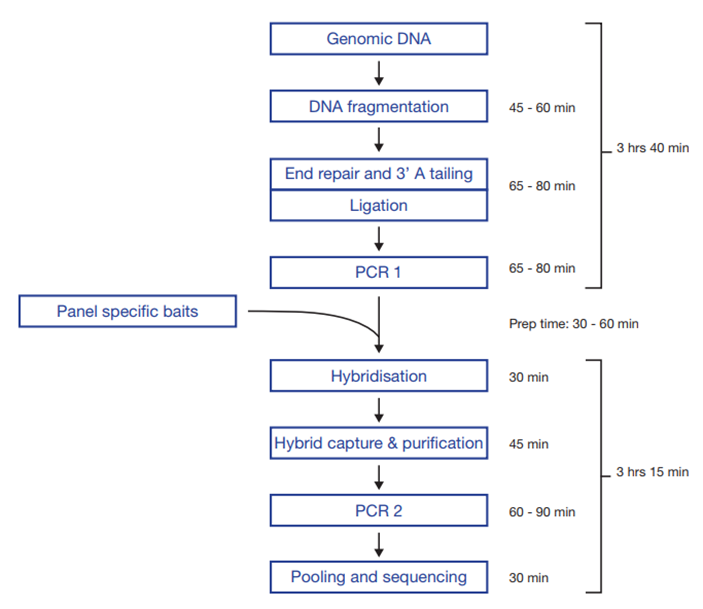 Figure 1: Workflow of SureSeq NGS library preparation, from DNA to sequencer.
Figure 1: Workflow of SureSeq NGS library preparation, from DNA to sequencer.
Panel content
- The SureSeq Core MPN Panel covers specific sites of MPN research relevance in 3 genes: JAK2 (V617F and exon 12), MPL (W515K/L/R/A and S505N) and CALR (exon 9).
Study design
- The SureSeq Core MPN Panel was validated using the JAK2 V617F Genotyping Sensitivity Panel provided by the National Institute for Biological Standards and Control (NIBSC) in order to confirm the lower levels of analytical detection and confidence.
- The panel was also used to confirm a broader set of variants in 14 research samples* containing variants for each of the targeted regions. Sequencing was conducted on a MiSeq® using a V2 300 bp cartridge (Illumina).
Results
Sensitive and reproducible variant detection even in heterogeneous samples
- Heterogeneous cancer samples pose significant challenges for researchers as alleles are likely to be present at a lower fraction than what would be expected for standard germline variants. Samples also typically contain a mixture of cancer and normal cells; moreover, cancer can consist of several molecularly distinct clones. In order to detect alleles that contribute only a small percentage to the reads at any locus, a highly uniform and sensitive enrichment is required.
- The SureSeq Core MPN Panel has been validated with samples from the NIBSC and have been shown to accurately detect alleles down to a 1% VAF in JAK2 (V617F) at a read depth of >1000x (Table 1).
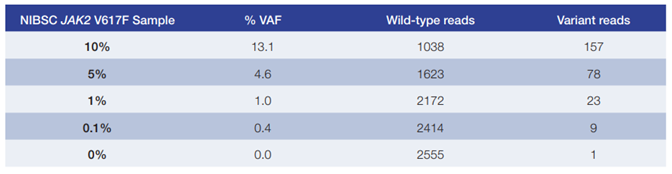 Table 1: Data generated from a 48 sample run on an Illumina MiSeq. The SureSeq Core MPN Panel permitted the detection of alleles at 1% VAF with high confidence.
Table 1: Data generated from a 48 sample run on an Illumina MiSeq. The SureSeq Core MPN Panel permitted the detection of alleles at 1% VAF with high confidence.
Evaluation of the Core SureSeq MPN Panel and enhanced LPK protocol with 14 research samples
- Data presented here are from 14 research samples (including 2 wild-type controls) that were processed using the enhanced LPK in combination with the SureSeq Core MPN panel.
- The range of variants and VAFs detected from 14 research samples included SNVs as well as 5 bp insertions in JAK2 (exon 12) and deletions of up to 52 bp in CALR (exon 9). No variants were identified in the control samples (Table 2).
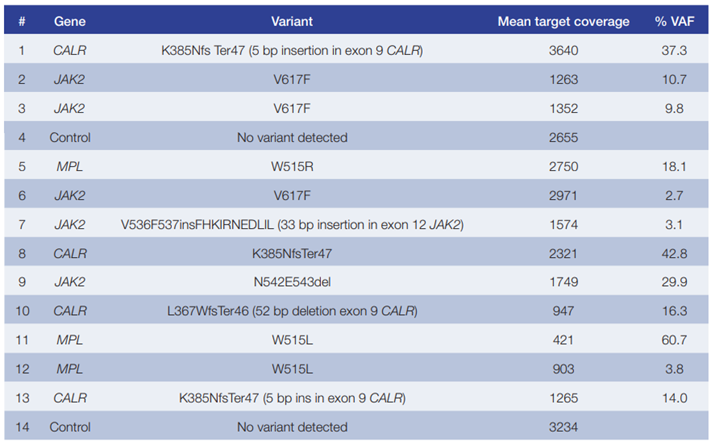 Table 2: Data generated using the SureSeq Core MPN Panel in combination with the enhanced LPK was 100% concordant with independent findings (National Genetics Reference Laboratory – Wessex, UK).
Table 2: Data generated using the SureSeq Core MPN Panel in combination with the enhanced LPK was 100% concordant with independent findings (National Genetics Reference Laboratory – Wessex, UK).
The SureSeq Core MPN panel in combination with the enhanced workflow is able to reliably detect single nucleotide variants (SNVs) as well as insertions (5 bp insertion in JAK2 exon 12 and CALR exon 9) and deletions (52 bp deletion CALR exon 9 (Figure 2) and 5 bp deletion exon 12 JAK2 (Figure 3)).
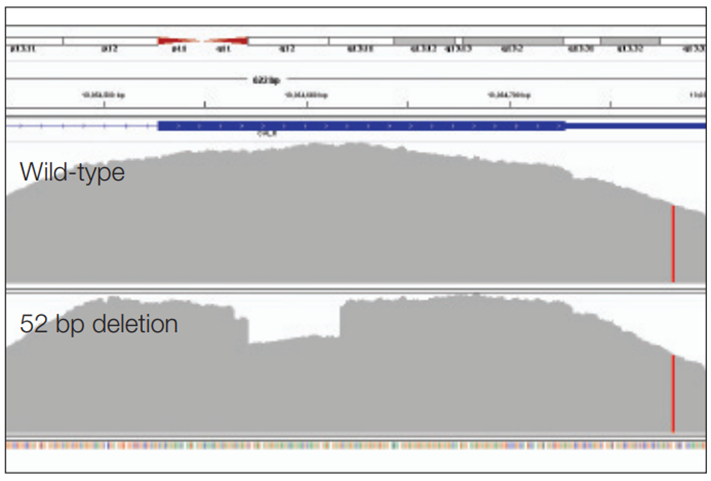 Figure 2: Detection of a 52 bp deletion (exon 9 CALR). Wild-type sample (top panel) is compared to a 52 bp somatic deletion (bottom panel).
Figure 2: Detection of a 52 bp deletion (exon 9 CALR). Wild-type sample (top panel) is compared to a 52 bp somatic deletion (bottom panel).
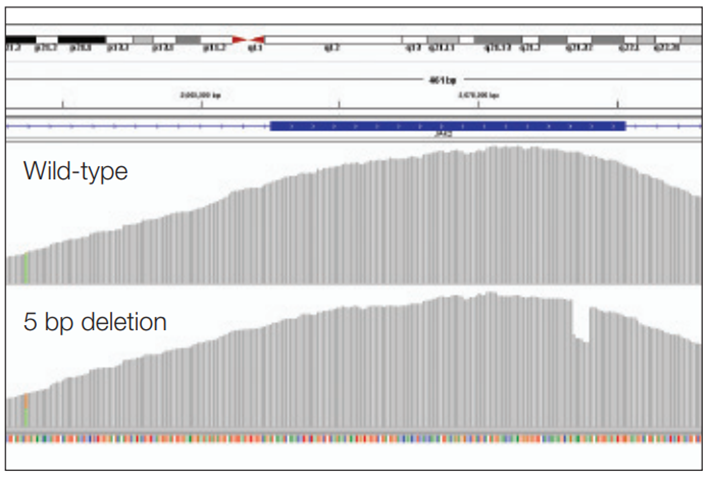 Figure 3: Detection of a 5 bp deletion (exon 12 JAK2). Wild-type sample (top panel) is compared to a 5 bp somatic deletion (bottom panel).
Figure 3: Detection of a 5 bp deletion (exon 12 JAK2). Wild-type sample (top panel) is compared to a 5 bp somatic deletion (bottom panel).
Conclusions
- We have successfully utilized the OGT 1-day hybridization-based SureSeq LPK protocol in combination with the SureSeq Core MPN Panel to reliably and confidently detect somatic SNVs by NGS down to a 1% VAF.
- The uniformity of coverage of this approach permitted the detection of key CALR and JAK2 variants (including 52 bp deletions and 5 bp insertions).
- This enhanced protocol incorporates an enzymatic fragmentation step which permits the high-throughput preparation of 48 samples from genomic DNA to sequencer in a 1-day workflow.
- To achieve >1000x de-duplicated depth (required for confident detection of 1% VAF), 48 samples can be reliably sequenced in a single MiSeq (V2 300 bp) run. This allows the generation of high quality data in a cost effective and timely manner.
Acknowledgements
*We would like to thank Prof. Nick Cross (National Genetics Reference Laboratories - Wessex, UK) for providing the validated research samples.
SureSeq: For Research Use Only; Not for Diagnostic Procedures.
 Figure 1: Workflow of SureSeq NGS library preparation, from DNA to sequencer.
Figure 1: Workflow of SureSeq NGS library preparation, from DNA to sequencer. Table 1: Data generated from a 48 sample run on an Illumina MiSeq. The SureSeq Core MPN Panel permitted the detection of alleles at 1% VAF with high confidence.
Table 1: Data generated from a 48 sample run on an Illumina MiSeq. The SureSeq Core MPN Panel permitted the detection of alleles at 1% VAF with high confidence. Table 2: Data generated using the SureSeq Core MPN Panel in combination with the enhanced LPK was 100% concordant with independent findings (National Genetics Reference Laboratory – Wessex, UK).
Table 2: Data generated using the SureSeq Core MPN Panel in combination with the enhanced LPK was 100% concordant with independent findings (National Genetics Reference Laboratory – Wessex, UK). Figure 2: Detection of a 52 bp deletion (exon 9 CALR). Wild-type sample (top panel) is compared to a 52 bp somatic deletion (bottom panel).
Figure 2: Detection of a 52 bp deletion (exon 9 CALR). Wild-type sample (top panel) is compared to a 52 bp somatic deletion (bottom panel). Figure 3: Detection of a 5 bp deletion (exon 12 JAK2). Wild-type sample (top panel) is compared to a 5 bp somatic deletion (bottom panel).
Figure 3: Detection of a 5 bp deletion (exon 12 JAK2). Wild-type sample (top panel) is compared to a 5 bp somatic deletion (bottom panel).