Want to download this as a PDF? Download now
DNA fragmentation is a crucial first step in the preparation of libraries for next generation sequencing (NGS). Mechanical shearing (sonication) is currently the gold-standard for fragmentation but it requires a significant upfront capital investment. OGT has evaluated an alternative method of fragmentation using the NEBNext® dsDNA Fragmentase®.
24 DNA samples isolated from whole blood were digested using NEBNext dsDNA Fragmentase (cat. no. M0348S). A second set of the same samples were sheared on a Covaris® Focused-ultrasonicator™. Libraries for sequencing were prepared using the SureSeq™ NGS Library Preparation Kit (cat. no. 500070) and enrichment was completed with the SureSeq™ Myeloid Panel (cat. no. 600075) (Figure 1).
All samples were sequenced on an Illumina MiSeq® using 150-base paired-end reads with the MiSeq Reagent Kit v2 300 cycle (cat. no. MS-102-2002). 24 samples were run on a MiSeq lane.
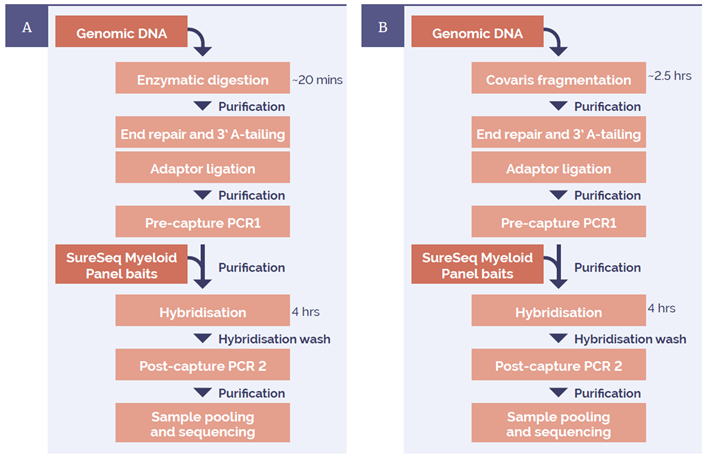 Figure 1: Library preparation workflow using (A) NEBNext enzymatic digestion and (B) Covaris mechanical shearing. Processing times: enzymatic digestion ~ 20 min total (5 min hands-on); Covaris method ~ 2.5 h total (hands-on). Times are based on processing 24 samples.
Figure 1: Library preparation workflow using (A) NEBNext enzymatic digestion and (B) Covaris mechanical shearing. Processing times: enzymatic digestion ~ 20 min total (5 min hands-on); Covaris method ~ 2.5 h total (hands-on). Times are based on processing 24 samples.
Enzymatic fragmentation can result in broader fragment size distribution, as shown in the Agilent® TapeStation traces below (Figure 2). This however did not affect the sequencing data quality.
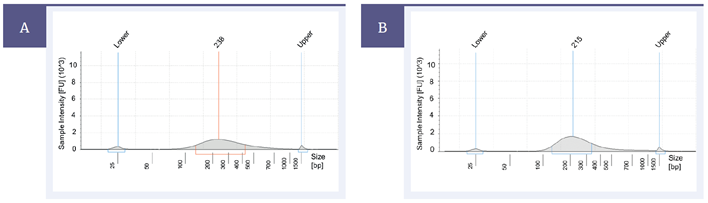 Figure 2: High quality genomic DNA fragmented by (A) NEBNext dsDNA Fragmentase for 20 min and(B) Covaris ultrasonicator (with fragmentation settings of 200 bp).
Figure 2: High quality genomic DNA fragmented by (A) NEBNext dsDNA Fragmentase for 20 min and(B) Covaris ultrasonicator (with fragmentation settings of 200 bp).
Both methods of fragmentation resulted in a very good mean target coverage as well as on-target rates (%OTR) meeting the technical specifications for the SureSeq Myeloid Panel (Figure 3). Uniformity of coverage is also unaffected by the method of DNA fragmentation (Figure 4).
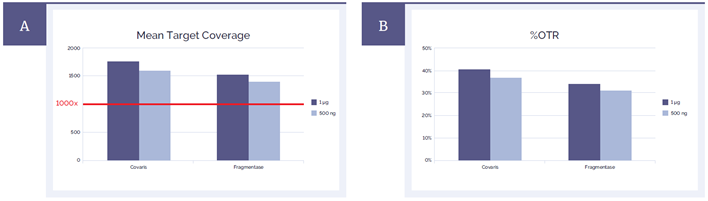 Figure 3: Sequencing metrics meet the technical specifications for the SureSeq Myeloid Panel. (A) Mean target coverage achieved using both methods exceeds 1000x required for detection of variants down to 1% minor allele fraction (MAF). (B) %OTR are on average 10% lower for the fragmentase method but are still within the acceptable range and very high for a panel of this size.
Figure 3: Sequencing metrics meet the technical specifications for the SureSeq Myeloid Panel. (A) Mean target coverage achieved using both methods exceeds 1000x required for detection of variants down to 1% minor allele fraction (MAF). (B) %OTR are on average 10% lower for the fragmentase method but are still within the acceptable range and very high for a panel of this size.
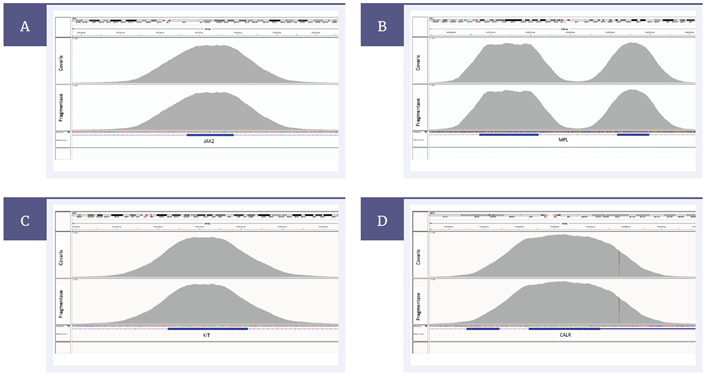 Figure 4: Coverage is very uniform for both methods. Shown here are selected exons of JAK2 (A) MPL (B) KIT (C) and CALR (D) genes. The top panel of each pair show samples prepared with Covaris and the bottom panels show samples prepared with the fragmentase method.
Figure 4: Coverage is very uniform for both methods. Shown here are selected exons of JAK2 (A) MPL (B) KIT (C) and CALR (D) genes. The top panel of each pair show samples prepared with Covaris and the bottom panels show samples prepared with the fragmentase method.
The recommended amount of input DNA for the SureSeq Myeloid Panel is 500 ng – 1 µg. Sometimes, however, the amount of material available for testing is limited. We have evaluated the effect of decreased amount of input DNA on the quality of NGS data with the fragmentase method. As expected, both mean target coverage and %OTR decrease with reduced amount of input DNA but even for as little as 100 ng the metrics still meet the specifications for this panel (Figure 5).
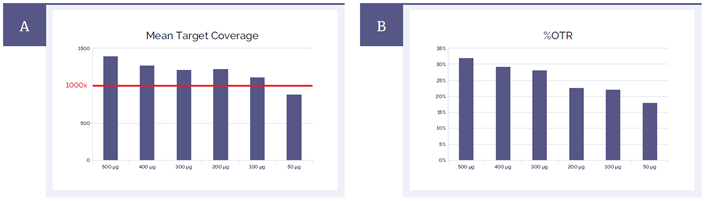 Figure 5: Effect of reduced amount of DNA input on mean target coverage (A) and %OTR (B).
Figure 5: Effect of reduced amount of DNA input on mean target coverage (A) and %OTR (B).
Despite a slight drop in the depth of coverage seen when reducing the DNA input, all sites of interest are covered to at least 1000x depth allowing confident detection of variants down to 1% minor allele fraction (MAF) (Figure 6).
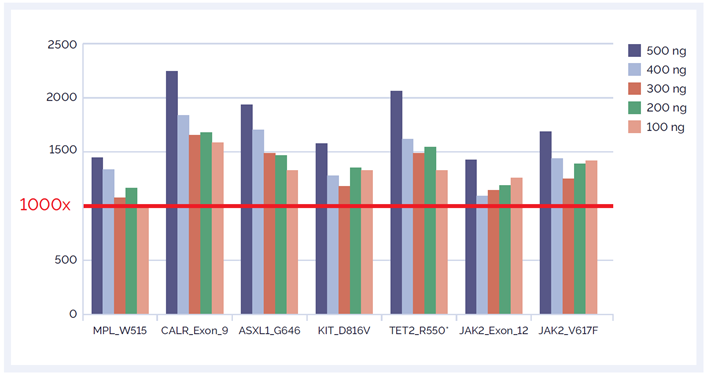 Figure 6: Coverage of the sites of interest on the SureSeq Myeloid Panel using various DNA input amounts with enzymatic fragmentation.
Figure 6: Coverage of the sites of interest on the SureSeq Myeloid Panel using various DNA input amounts with enzymatic fragmentation.
Uniformity of coverage across the panel is another important NGS metric. Gaps in coverage or presence of regions where sequencing depth is significantly lower compared to the mean depth can results in false negative calls. To confidently call variants, particularly in heterogeneous cancer samples where variants are often present at low fractions, it is essential that all regions targeted by the panel are covered uniformly. We have shown that reducing the amount of input DNA has no effect on the uniformity of coverage: 100% of bases are covered to at least 20% of the mean and >98% of bases are covered to at least 50% of the mean (Figure 7).
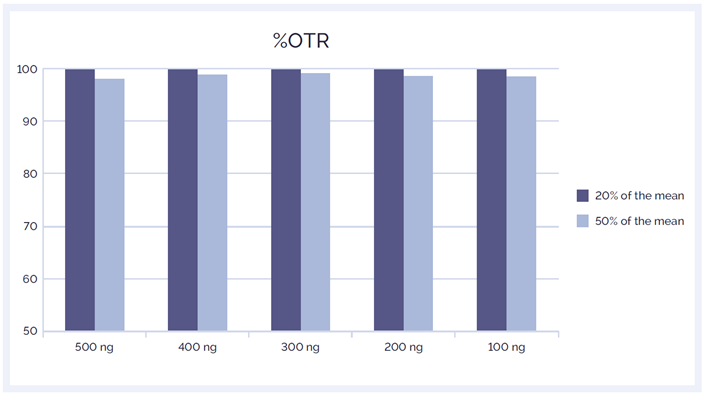 Figure 7: The SureSeq Myeloid Panel delivers excellent uniformity of coverage regardless of the amount of input DNA.
Figure 7: The SureSeq Myeloid Panel delivers excellent uniformity of coverage regardless of the amount of input DNA.
Enzymatic digestion using NEBNext dsDNA Fragmentase provides a fast and efficient alternative to mechanical shearing. Metrics including on-target rates and mean target coverage remain comparable to those achieved using the standard Covaris method.
Additionally, the fragmentase method allows a reduced amount of input DNA to be used without affecting the panel’s sensitivity or uniformity of coverage. High-quality NGS data can be obtained using as little as 100 ng of good-quality DNA isolated from whole blood. Using enzymatic digestion instead of sonication can speed up the process of library preparation and save up to 2 h of hands-on time.
SureSeq™: For Research Use Only; Not for Diagnostic Procedures.This document and its contents are © Oxford Gene Technology IP Limited – 2021. All rights reserved. OGT™ and SureSeq™ (Oxford Gene Technology); Agilent® (Agilent Technologies Inc.); MiSeq® (Illumina Inc.); Covaris® and Focused-ultrasonicator™ (Covaris, Inc.); NEBNext® and Fragmentase® (New England Biolabs, Inc.). The SureSeq NGS Library Preparation Kit was jointly developed between Oxford Gene Technology and Bioline Reagents Limited. Dynabeads is a trademark of Thermo Fisher Scientific and AMPure® is a registered trademark of Beckman Coulter Inc.
