One of the greatest challenges for laboratories running next generation sequencing (NGS) panels is to ensure high quality, reproducible results whilst sustaining high sample throughput in a timely fashion. Automation enables standardization of Oxford Gene Technology’s (OGT) SureSeq™ NGS library preparation, hybridisation and washing for up to 96 samples in a single batch, improving laboratory productivity. In this application note, an Agilent Bravo® A Automated Liquid Handling Platform was configured to run the SureSeq NGS library preparation protocol. The results demonstrate marked improvement not only in hands-on-time, but also a number of quality metrics, in particular, reduced variance in % on-target reads when compared to manual processing.
Varying amounts of good quality genomic DNA were used as templates. Samples were fragmented using NEBNext® dsDNA Fragmentase® (New England Biolabs Inc. MO348S), libraries were prepared with an Agilent Bravo A Automated Liquid Handling Platform using the SureSeq NGS Library Preparation Kit and hybridized to the SureSeq myPanel™ Custom Myeloid Panel – 49 gene Plus (602017). Resulting libraries were grouped into 16 sample batches and sequenced on an Illumina™ MiSeq (2 x 150 bp v2 cartridge MS-102-2002). Sequencing data was processed using OGT’s SureSeq Interpret Software.
In hybridization-based target enrichment, genomic DNA is first randomly sheared into 150 – 250 bp fragments by mechanical or enzymatic fragmentation. OGT offers scripts compatible with both mechanical and enzymatic fragmentation workflows, producing highly reproducible results across a range of starting input amounts (Figure 1).
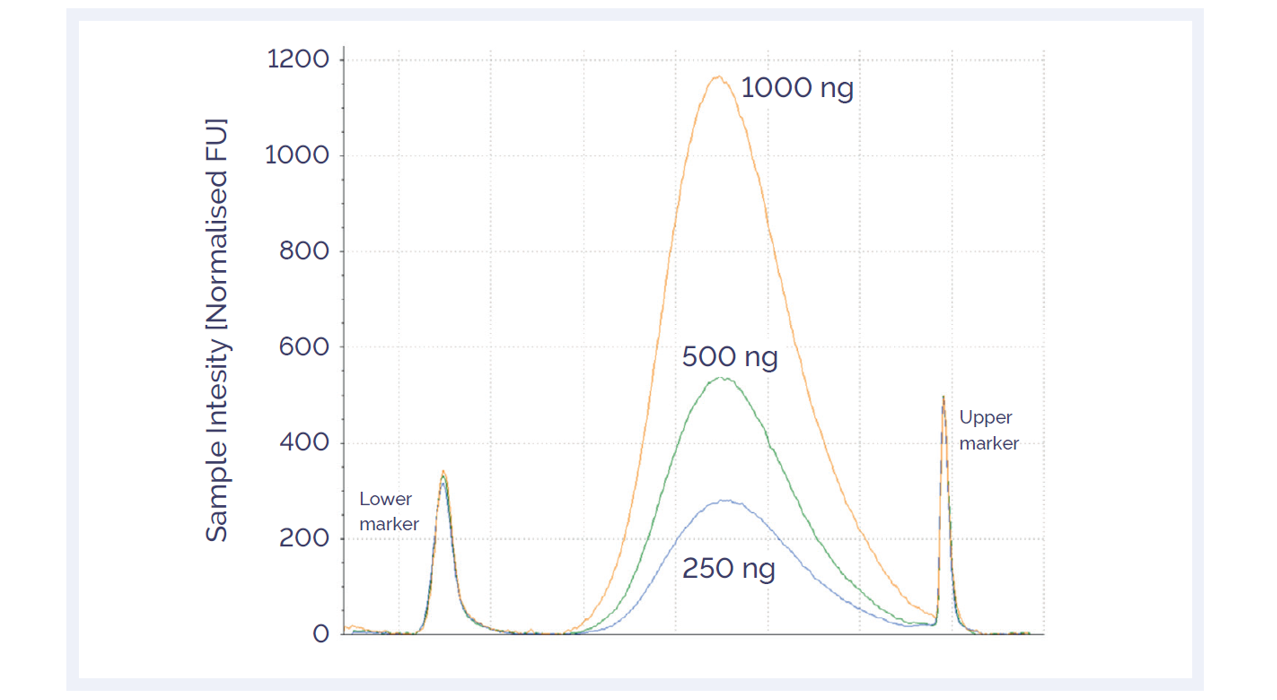
Figure 1: Agilent TapeStation™ trace of enzymatically sheared good quality DNA. Master mix preparation and sample handling, including AMPure® purification, were performed on the Bravo instrument. The fragment sizes generated ranged from 150–250 bp regardless of amount of starting DNA. The recommended fragment size for the SureSeq NGS range is 150–250 bp.

Reducing the labor intensity of any step in any laboratory workflow is highly desirable. As the hybridization-based enrichment workflow contains routine steps such as reagent/sample transfers and bead-based purifications, it is highly amenable to automation and the benefits that brings compared to manual processing, for example in the reduction in the amount of hands-on time required (Figure 2).
NGS hybridization and washing involves a significant number of pipetting steps which, when performed manually, can introduce high levels of variability. Apart from the requirement for highly skilled personnel, large numbers of manual steps can result in user fatigue, and in some cases errors. Automation offers a solution which can minimize or eliminate these issues, in particular that of increased experimental variability. Samples processed by OGT’s scripts are highly reproducible across a range of starting inputs (Figure 3) and show a reduction in variability compared to manually processed samples (Figure 4).
Hybridization-based enrichment protocols often require users to concentrate their unenriched samples using a SpeedVac® or similar vacuum concentration device; this adds time to the total workflow. OGT have developed a bead-based method to concentrate unenriched samples, this alternative workflow is compatible with the Agilent Bravo and delivers highly reproducible data across a range of starting DNA inputs (Figure 5).
Samples processed by the Agilent Bravo maintain and sometimes improve upon the excellent levels of uniformity seen over a variety of targets (Figures 6a and 6b) including difficult targets such as the FLT3 ITD region as shown in Figure 7.
The Bravo A Automated Liquid Handling Platform was successfully set up to run the SureSeq NGS library preparation workflow. The results showed improved reproducibility with a threefold reduction in the coefficient of variation in % on-target reads of when compared to manual processing, whilst hands on time was reduced by a third.
The ultimate goal of any sequencing assay is to discover all variants present; uniformity of enrichment means that all regions are represented more equally. With this in mind, the excellent reproducibility of mean target coverage and coverage uniformity exhibited by the hybridization-based enrichment of SureSeq panels in conjunction with the Bravo Platform facilitates the reliable detection of low-frequency somatic variants. With automated preparation allowing the processing of up to 96 samples at any one time, it offers excellent reliability and reproducibility with easy scale up of laboratory throughput.
SureSeq™: For Research Use Only; Not for Diagnostic Procedures. The SureSeq NGS Library Preparation Kit was jointly developed between OGT and Bioline Reagents Limited. SpeedVac® is a registered trademark of Thermo Fisher Scientific, Covaris™ is a registered trademark of Covaris, Inc., Bravo® and TapeStation™ are registered trademarks of Agilent Technologies, Inc., NEBNext® and Fragmentase® (New England BioLabs, Inc.) are registered trademarks of New England Biolabs, Inc. AMPure® is a registered trademark of Beckman Coulter, Inc.
Call +44 (0)1865 856800 Email contact@ogt.com
Send us a message and we will get back to you