Wojtaszewska Marzena1, Szarawarska Marta1, Skoczylas Tomasz1, Markiewicz Mirosław1, 2
1 Department of Hematology, Clinical University Hospital in Rzeszow, Poland
2 Department of Hematology, Institute of Medical Sciences, College of Medical Sciences, Rzeszow University, Poland
The need for better and faster molecular risk stratification has continued to emerge alongside the development of novel treatment regimens for chronic lymphocytic leukemia (CLL). Traditionally, comprehensive genetic testing in CLL requires multiple laborious and costly diagnostic strategies. Traditionally, actionable copy number alterations (CNAs) are tested by cytogenetic methods. Typically, these CNAs are heterogeneous in size and the detection of CNAs smaller than 1Mbp by FISH is often challenging for the cytogeneticist. The application of next-generation sequencing may serve to help reduce the burden of issues associated with CLL testing.
In this study we analyze the concordance of CNA size and architecture as well as the sensitivity of small variant detection using OGT’s SureSeq™ CLL+CNV V3 Panel, FISH and Nextera® sequencing.
23 pathogenic variants in TP53 were detected with allele burden (VAF) of 1-81%. 7 variants in SF3B1 were detected (VAF ranged 1-48%). All were confirmed by the orthogonal method. Two resistance mutations in BTK were also detected by OGT’s SureSeq CLL+CNV V3 Panel in patients resistant to the BTK inhibitors. Moreover, BRAF and KRAS pathogenic variants were also detected and confirmed by Nextera sequencing2.
Within the declared detection limit of the NGS panel, there were 9 deletions in 11q, 10 losses of 17p, two trisomies and one partial duplication of 12q and 11 deletion of 13q. The size of aberrations ranged from 298kbp to full chromosomes. All aberrations were confirmed by FISH. Deletions smaller than 1Mbp were described in FISH as “dim”, indicating correct hybridization pattern was present with different signal intensities (example shown in Fig.1.)
In case of 13q deletions both FISH and NGS concordantly described partial losses and compound deletions of DLEU and/or RB1 loci. In 4 instances 13q deletion was further characterized as clonally heterogeneous, manifesting in two distinct hybridization patterns in FISH and in differences of logR in NGS (example shown in Fig. 2.).
Samples harboring CNAs with less than 20% aberration burden in FISH were not detected by the SureSeq™ CLL+CNV V3 Panel, as they were below its detection limit.
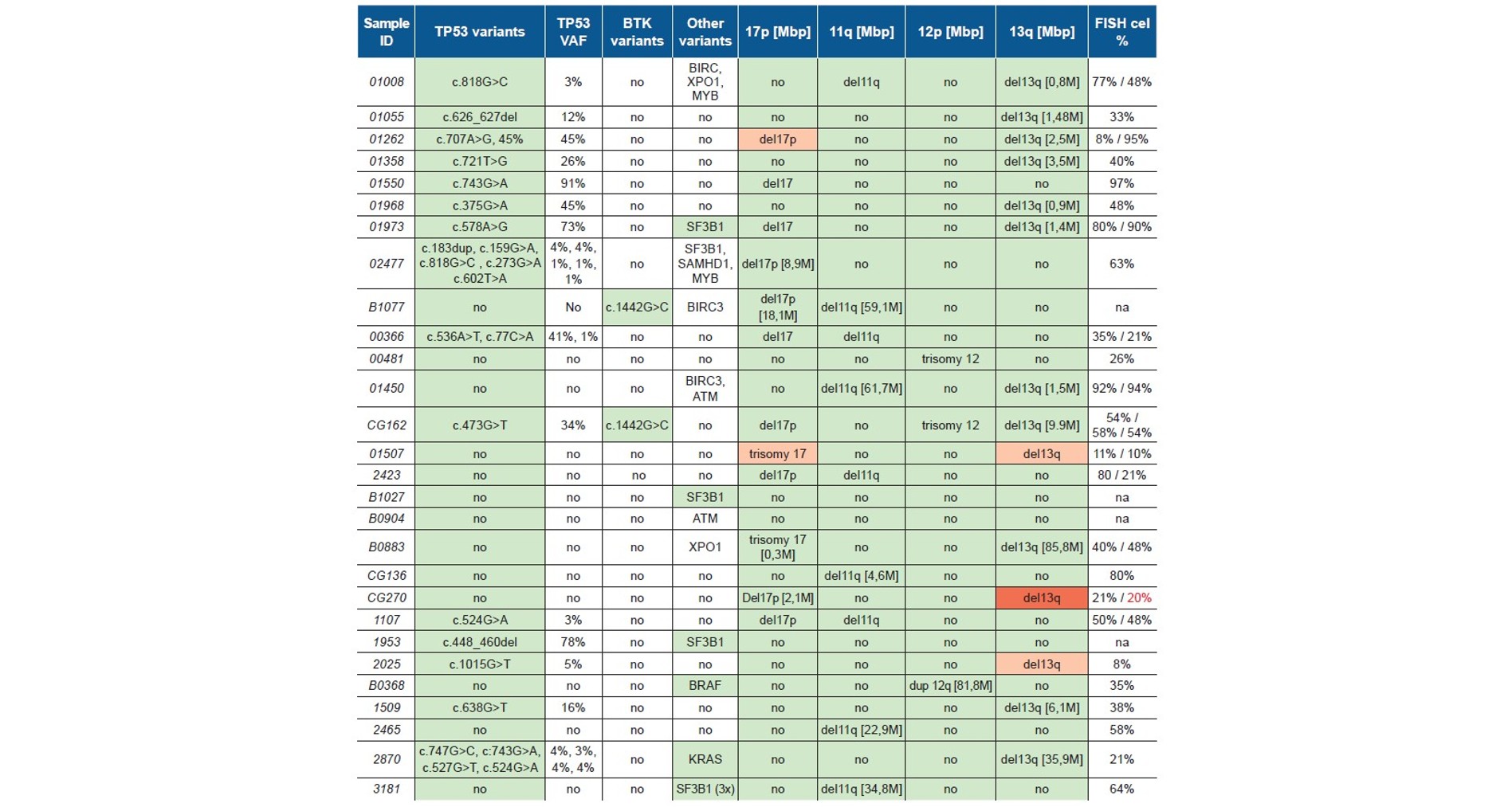
Table 1: CNVs and small pathogenic variants detected by OGT’s SureSeq™ CLL+CNV V3 Panel. The variants in green were confirmed by orthogonal methods: Nextera sequencing (SNV/indel) or FISH (CNVs). In orange are variants that were detected by orthogonal method but were not confirmed by OGT’s SureSeq CLL + CNV V3 Panel as they were below its detection limit. In red are variants detected by FISH, but not detected by OGT’s SureSeq CLL + CNV V3 Panel as they were below its detection limit.

We demonstrate that hybrid-capture next generation sequencing is an efficient tool combining small variant and CNA detection in clinical CLL samples. The actionable mutations of at least 2,5% VAF were called, as well as all of disease associated clonal copy number alterations within the methods detection limit (20%). Our findings confirm that applicability of NGS panels in CLL diagnostics3. The benefit of NGS testing is its higher resolution, enabling reliable detection of < 1Mbp alterations which may be overlooked by the human eye during FISH analysis.
The corresponding author of the poster received research support in the form of reagents and travel reimbursement from Oxford Gene Technology Ltd., Unit 5, Oxford Technology Park, 4A Technology Drive, Kidlington, Oxfordshire, OX5 1GN, UK.
This presentation is intended for educational purposes only and does not replace independent professional judgment. Statements of fact and opinions expressed are those of the presenters individually and, unless expressly stated to the contrary, are not the opinion or position of the Oxford Gene Technology Group (OGT). OGT does not endorse or approve, and assumes no responsibility for, the content, accuracy or completeness of the information presented.
SureSeq™: For Research Use Only; Not for Use in Diagnostic Procedures.
Call +44 (0)1865 856800 Email contact@ogt.com
Send us a message and we will get back to you