Want to download this as a PDF? Download now
Lyudmila Georgieva, Ezam Uddin, Aysel Heckel, Jacqueline Chan, James Reid, Jolyon Holdstock, David Cook and Graham Speight
Karyotyping, FISH, RT-PCR and microarrays are currently considered to be the gold standard techniques for structural variant discovery and detection. However, there is a desire to combine analysis of large structural alterations such as translocations alongside smaller mutations such as SNVs and indels. With development of newer technologies such as NGS for DNA- and RNA-sequencing, simultaneous discovery of multiple mutation types is now possible, enabling development of more comprehensive assays.
Chronic myeloid leukemia (CML) is a myeloproliferative neoplasm with incidence of 1 to 2 per 100,000 and constitutes 15-20 % of adult leukemias1. CML is characterized by the Philadelphia chromosome (Ph), resulting from the t(9;22)(q34;q11) balanced reciprocal translocation. The translocation generates the BCR-ABL1 fusion gene encoding the BCR-ABL1 protein with constitutive kinase and oncogenic activity1,2. The breakpoints in the ABL1 gene lie in 90 kb long intron 1, upstream of the ABL1 tyrosine kinase domains encoded in exons 2 to 11. The breakpoints within BCR are mapped to a 3.1 kb area spanning exons 13 to 15, the major breakpoint cluster region (M-bcr), found in 90% of CML and 20 to 30% of cases with Ph-positive B-cell acute lymphoblastic leukemia (Ph+ B-ALL)3.
In this study, we tested the capability of a SureSeq myPanel™ NGS Custom Cancer Panel to detect known t(9;22)(q34;q11) translocations.
The SureSeq™ hybridization-based approach was used throughout this study; the workflow of this is outlined in Figure 1.
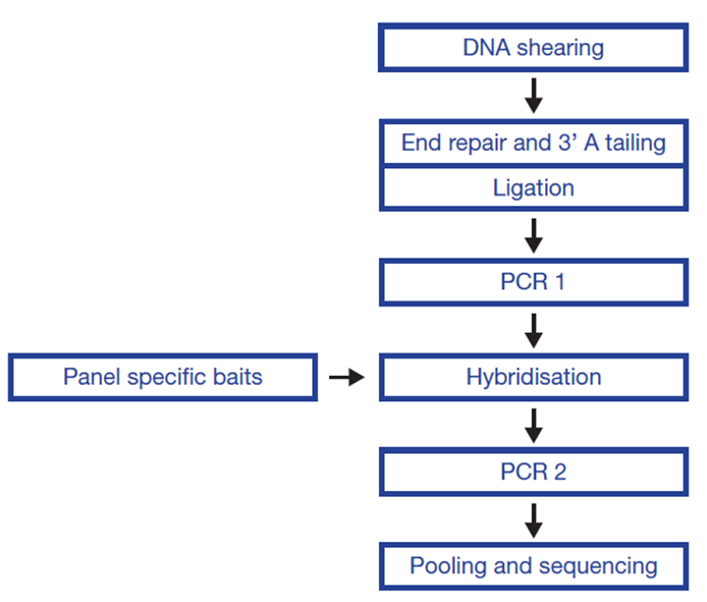 Figure 1: OGT SureSeq workflow. The SureSeq workflow allows users to go from extracted DNA to sequencer in 1.5 days with minimal handling time.
Figure 1: OGT SureSeq workflow. The SureSeq workflow allows users to go from extracted DNA to sequencer in 1.5 days with minimal handling time.
We utilized a SureSeq myPanel NGS Custom panel and associated library preparation kit* to determine whether this approach can be used for detection of the t(9;22)(q34;q11) translocation.
We used a hybridization-based enrichment approach to test characterized DNA samples, CML cell lines K562, KU-812, MOLM-1 and JK-1, and 3 research samples** with breakpoints occurring across multiple positions in the major breakpoint area of BCR (exons 13-15) (Figure 2).
To mimic the BCR-ABL1 translocation with different frequencies we made a serial dilution of cell line K562 in order to create translocation carriers with frequency range 0 - 100%.
Sequencing was conducted using a V2 300 bp cartridge (Illumina).
Data was analysed using OGT’s proprietary translocation detection software.
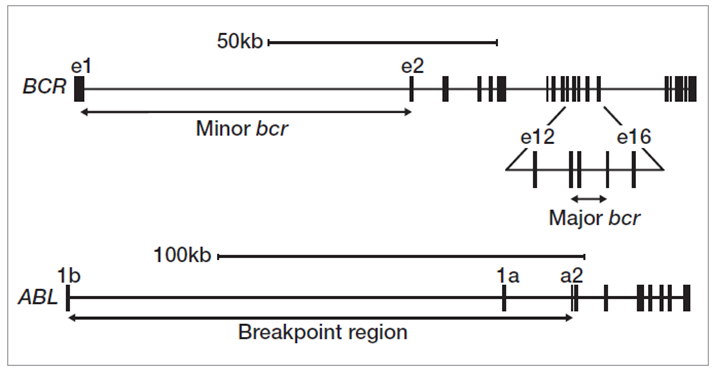 Figure 2: Translocation breakpoint clusters in BCR and ABL1 genes.
Figure 2: Translocation breakpoint clusters in BCR and ABL1 genes.
We have achieved high depth (>1000x) and uniformity of coverage across the targeted regions which enabled successful detection of all translocation events.
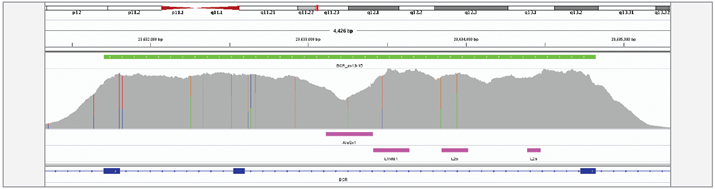 Figure 3: Excellent uniformity of coverage of the BCR, exon 13-15 region, generated with OGT SureSeq protocol averaging ~1500x deduplicated coverage. Depth of coverage per base (grey). Target region (green). Repeat regions (pink).
Figure 3: Excellent uniformity of coverage of the BCR, exon 13-15 region, generated with OGT SureSeq protocol averaging ~1500x deduplicated coverage. Depth of coverage per base (grey). Target region (green). Repeat regions (pink).
Using the OGT workflow we were able to reliably detect BCR-ABL1 translocations in 4 CML cell lines. For all cell lines, predicted breakpoints were found to match the coordinates reported in the literature4 (Table 1, Figure 4).
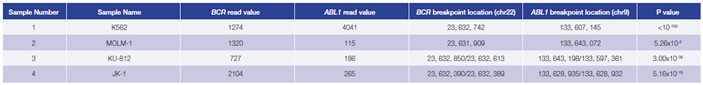 Table 1: Accurate detection of BCR-ABL1 translocations by early stage software (in development). Read values are a formulation of read depth, not true read depth.
Table 1: Accurate detection of BCR-ABL1 translocations by early stage software (in development). Read values are a formulation of read depth, not true read depth.
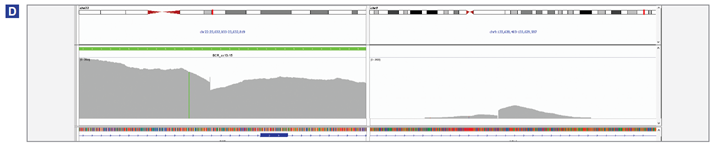
Figure 4 (below): BCR-ABL1 translocation detection in K562 (A), MOLM-1 (B) , KU-812 (C) and JK-1 (D) cell lines.
 (A) Breakpoint location BCR chr22 : 23,632,742, ABL1 breakpoint location chr9 : 133,607,145.
(A) Breakpoint location BCR chr22 : 23,632,742, ABL1 breakpoint location chr9 : 133,607,145.
 (B) Breakpoint location BCR chr22 : 23,631,909, ABL1 breakpoint location chr9 : 133,597,361.
(B) Breakpoint location BCR chr22 : 23,631,909, ABL1 breakpoint location chr9 : 133,597,361.
 (C) Both BCR-ABL1 and ABL1-BCR translocations were detected. BCR/ABL1 : chr22 : 23,632,850 / chr9 : 133,643,198; ABL1/BCR: chr9 : 133,643,072 / chr22 : 23,632,613.
(C) Both BCR-ABL1 and ABL1-BCR translocations were detected. BCR/ABL1 : chr22 : 23,632,850 / chr9 : 133,643,198; ABL1/BCR: chr9 : 133,643,072 / chr22 : 23,632,613.
 (D) Both BCR-ABL1 and ABL1-BCR translocations were detected. BCR/ABL1 : chr22 : 23,632,390 / chr9 : 133,628,935; ABL1/BCR: chr9 :133,628,932 / chr22 : 23,632,389.
(D) Both BCR-ABL1 and ABL1-BCR translocations were detected. BCR/ABL1 : chr22 : 23,632,390 / chr9 : 133,628,935; ABL1/BCR: chr9 :133,628,932 / chr22 : 23,632,389.
This data (Figure 5, Table 2) is based on a serial dilution of DNA from K562 cell line in order to create BCR-ABL1 translocation carriers with frequencies of 0 – 100%.
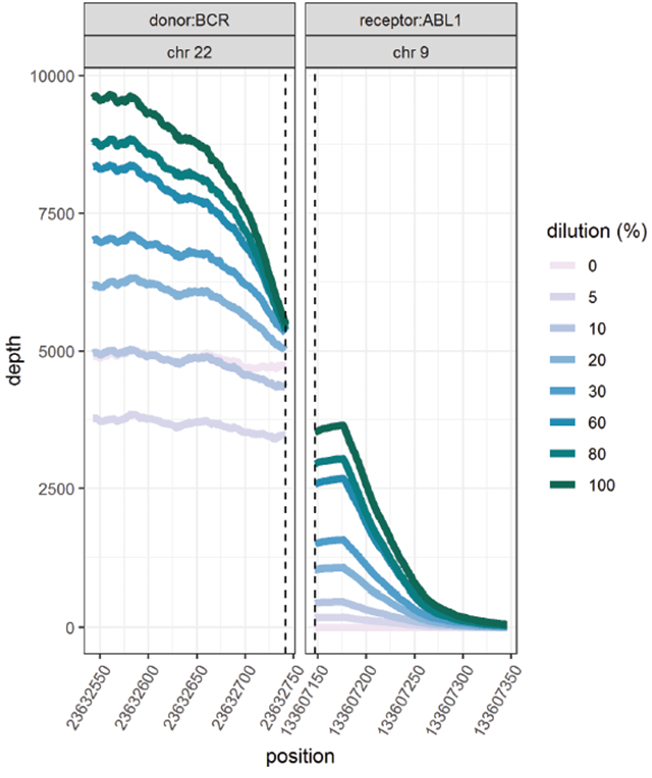
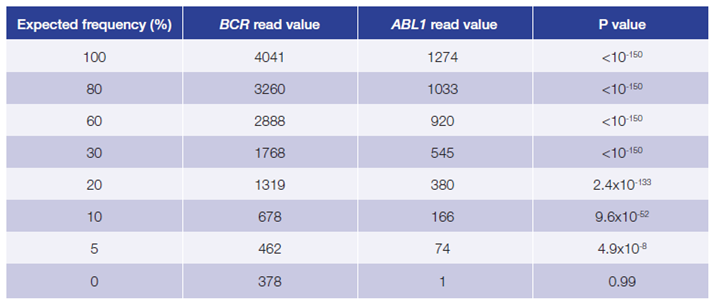 Figure 5, Table 2: Accurate and reproducible detection of BCR-ABL1 translocation in K562 cell line (BCR breakpoint location chr22:23632742, ABL1 breakpoint location chr9:133607147) with a range of frequencies. Read values are a formulation of read depth, not true read depth.
Figure 5, Table 2: Accurate and reproducible detection of BCR-ABL1 translocation in K562 cell line (BCR breakpoint location chr22:23632742, ABL1 breakpoint location chr9:133607147) with a range of frequencies. Read values are a formulation of read depth, not true read depth.
Data presented here (Table 3) are from 3 research samples from carriers of a BCR-ABL1
translocation generated using the OGT workflow. For all samples the predicted breakpoints were found to be 100% concordant with independent findings (West Midlands Regional Genetics Laboratory, Birmingham – UK).
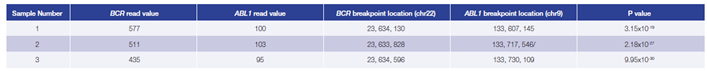 Table 3: Accurate detection of BCR-ABL1 translocations. Using the OGT workflow we were able to reliably detect BCR-ABL1 translocations with different breakpoint locations and frequency in all samples by early stage software (in development). Read values are a formulation of read depth, not true read depth.
Table 3: Accurate detection of BCR-ABL1 translocations. Using the OGT workflow we were able to reliably detect BCR-ABL1 translocations with different breakpoint locations and frequency in all samples by early stage software (in development). Read values are a formulation of read depth, not true read depth.
**Samples kindly provided by West Midlands Regional Genetic Laboratory - Birmingham, UK.
*The SureSeq NGS Library Preparation Kit was jointly developed between OGT and Bioline Reagents Limited.
SureSeq: For Research Use Only; Not for Diagnostic Procedures.
