Fluorescence in situ hybridization, better known as FISH, is a molecular cytogenetics technique that harnesses fluorescent probes that bind to specific nucleic acid sequences. FISH allows researchers to visualize and map genetic material on a chromosome to identify specific genes or portions of genes.
Since its discovery in the 1970’s1, FISH has revolutionized cytogenetics, ushering in a new era of molecular research. Of particular importance is its contribution to the field of oncology, where FISH has become an indispensable tool in the diagnosis, prognosis, and surveillance of many cancers.
Here, we discuss the important applications of FISH in cancer research and highlight its role as the gold standard for cancer diagnostics. Then, we will explore how alternative cytogenetic technologies may serve to complement its application.
Over half a century has passed since pioneering researchers realized that in situ hybridization (ISH) could be used to identify the positions of specific DNA sequences on chromosomes2.
Molecular cytogenetic techniques that harness ISH use labelled DNA or RNA sequences (probes). The probe hybridizes to its complementary (target) sequence in a biological sample. Depending on the label, hybridized sequences can be detected by methods such as autoradiography, fluorescence microscopy, or chromogenic detection.
In a landmark paper, Gall and Pardue (1969)2 were the first to describe how radioactive copies of a ribosomal DNA sequence could be used to detect complementary DNA sequences in the nucleus of a frog egg.
Within a decade, researchers had replaced radioactive labels with fluorescent labels in hybridization probes1, marking the transition from ISH to FISH. The rudimentary principles and steps that remain are similar to ISH where (Figure 1):
However, unlike previous ISH approaches, the use of FISH probes allow straight-forward detection of target sequences using a fluorescent microscope.
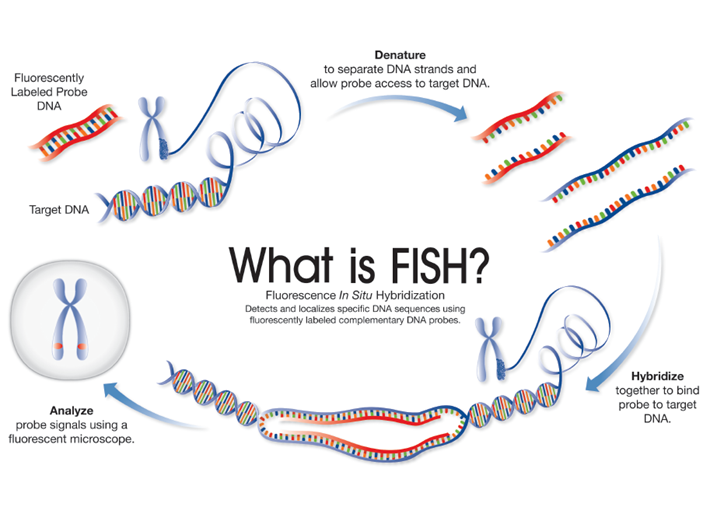
Figure 1: FISH and cytogenetics - schematic depicting each step involved in FISH
Since their discovery, FISH assays have become the gold standard for the diagnosis, prognosis, and surveillance of many cancer types – and for a good reason. In cancer diagnostics, FISH assays have been shown to increase detection capacity significantly compared to conventional cytogenetics.
For instance, acute myeloid leukemia (AML) is a hematologic neoplasm in which 33-50% of positive specimens have a normal karyotype3. Using FISH in cytogenetic laboratories allows for high-resolution analysis of recurrent structural chromosomal rearrangements recognized by the World Health Organization (WHO) as distinct disease entities within AML.
Find out more about CytoCell hematology FISH probes for AML/MDS
An additional advantage of FISH is its ability to offer rapid diagnostic results – a criticality when clinical decisions are pressing.
FISH can also be used as a prognostic tool. In other words, it can be used to make predictions around the likely course and outcome of a disease. For instance, loss of the TP53 (tumor protein p53) may be indicative of a poor outcome and is often seen as a marker of AML and MDS progression or secondary disease5. Appropriate FISH probes, such as the CytoCell Del(20q) Deletion and the CytoCell P53 (TP53) Deletion, can be used to detect these abnormalities and help inform clinical decisions.
Find out more about CytoCell FDA-cleared probes for AML and MDS
Chromosomal abnormalities specific to certain cancers can give a good indication of treatment response and outcome. For example, the HER2 (ERBB2) Human Epidermal growth factor Receptor 2 gene, located at chromosome band 17q12, is activated and amplified in 20-30% of breast cancers. Patients harboring this gene amplification may respond to HER2 inhibitors such as trastuzumab (Herceptin), and consequently, identified by FISH can inform treatment decisions6.
Finally, FISH assays can be used as surveillance tools to follow the effectiveness of treatment. In simple terms, an effective treatment will lead to a reduction in the number of abnormal cells seen in a patient sample, while continued detection of chromosomal abnormalities may indicate the presence of residual disease.
While FISH represents a robust, easily applicable technique with short turnaround times (TAT), it does have some limitations. For example, designing new assays can be a time-consuming process, where each stage of the protocol is probe- and sample-specific7. As such, multiple probes and precious clinical samples may be required to optimize clinically-validated products.
To address this, companies such as OGT offer an innovative custom probe design and manufacture service. By utilizing CytoCell’s proprietary BAC clone collection of more than 220,000 clones, OGT can produce fully quality-assured custom FISH probes for virtually any sequence in the entire human genome.
With this wide availability of FISH probes comes a wealth of diagnostic and research opportunities; including the detection of small chromosomal changes that are too small to be detected by conventional methods. Indeed, chromosomal microdeletions, that are usually <5Mb, evade identification by light microscopy and karyotyping but can be detected by FISH.
Microdeletions, or sub-microscopic deletions, are classically associated with constitutional diseases however, they can also be seen as acquired abnormalities in some cancer types.
With companies such as OGT expanding probe availability and helping to streamline the assay development and optimization workflow, it is certain that FISH is here to stay. Nevertheless, exploring other technologies, such as NGS, can offer additional or complementary insight in the field of cancer research.
While FISH remains an indispensable tool in cancer research, more modern techniques can be used to reveal unique and valuable information that can support FISH in cancer diagnostics, prognostics, and surveillance. Over recent years, one of the most notable developments in genetic disease research is the use of next-generation sequencing (NGS) technology.
NGS can capture a large amount of genomic information about cancer. In turn, it provides an opportunity to cast a wide net when many diverse mutations are indicative of disease.
Since FISH and NGS provide different advantages and have the potential to deliver unique genomic information, their utility is not mutually exclusive. For example FISH can provide rapid results and provides a valuable technique for the first round of testing and in time-critical scenarios. When negative FISH results demand further testing, NGS offers an approach to profile a larger genetic space.
Combining FISH with other techniques such as NGS, is just one approach to overcoming some of its limitations. However, advances in FISH technology, including automated workflows and streamlined probe design and manufacture, are serving to strengthen its position as the gold standard for detecting many genetic aberrations in cancer.
Whether your needs are off-the-shelf or custom, OGT can provide high-quality FISH probes optimized for your application. Discover a wide range of CytoCell FISH probes optimized for hematological malignancies and the assessment of genetic aberrations in solid tumor samples. If you can’t find what you’re looking for, at OGT we’re here to help with our CytoCell myProbes® - a custom FISH probe design and manufacture service.

Carmen Vokaty, Assistant-Chief Medical Technologist in Cytogenetics at the McGill University Health Centre, details how her lab incorporated OGT’s CytoCell® Tissue Pretreatment kit into its workflow after struggling with a homebrew solution.
Read
Dr Jennie Thurston, Director of Cytogenetics, Carolinas Pathology Group, Atrium Health, explains how OGT's CytoCell® FISH probes were seamlessly incorporated into the automated workflow for fast, high quality results.
Read
Megumi Hada, PhD, Senior Research Scientist, RaISE, Prairie View, Texas A&M. answers our questions on space radiation, how it affects astronaut health, and what her research can teach us about the biological effects of radiation here on Earth.
Read
Dr. Theresa C. Brown, Tulane University School of Medicine, answer questions around her AGT 2017 presentation 'FISHing with the real-life laboratory experts.'
Read