Want to download this as a PDF? Download now
 Genetic diseases are complex and require an analytical approach that is both comprehensive and flexible. In this white paper, genetic scientists Dr Tracey Lewis and Dr Emily Farrow discuss how customisable, exon focused array designs complement next generation sequencing (NGS) for clinical genetic research.
Genetic diseases are complex and require an analytical approach that is both comprehensive and flexible. In this white paper, genetic scientists Dr Tracey Lewis and Dr Emily Farrow discuss how customisable, exon focused array designs complement next generation sequencing (NGS) for clinical genetic research.
In light of recent advances in DNA analysis technologies, we are becoming ever more aware of the underlying complexity of genetic disorders. It is becoming increasingly evident that such disorders are rarely caused by an isolated mutation within a single gene, and one or many genes can contribute to one or multiple disorders. Moreover, causal aberrations vary widely from larger copy number variations (CNVs), down to single-point mutations. It is therefore vital to employ a suite of assays for assessing the entire scope of mutations within the exons of many different medically-relevant genes, creating a true reflection of the underlying genetic landscape of the ‘medical exome’.
Research scientist Dr Tracey Lewis from the Associated Regional and University Pathologists (ARUP) laboratories, and Director of Laboratory Operations at Children’s Mercy Hospital, Dr Emily Farrow, here explain their approaches to examining the medical exome. Outlining a comprehensive testing strategy, they discuss how the latest medically-focused microarray technology is applied alongside next generation sequencing (NGS) to provide unprecedented insights into causal genetic variation.
In order to accurately identify CNVs across all medically-relevant genes, there was a clear need throughout the medical research community for a focused array combining the latest genomic content with high-resolution exon-level probe design. Designing this type of array is challenging for two main reasons. Firstly, research is advancing at an incredible rate, as Dr Farrow explains: “The greatest challenge to the array is keeping up with the medically-relevant genes, as the list is updated daily.” Secondly, validating the probes against these newly discovered regions of the genome consumes both time and resources.
OGT has responded to this need with its CytoSure® Medical Research Exome array (MREA), developed in collaboration with Professor Madhuri Hegde* from Emory Genetics Laboratory – world leaders in genetic analysis and rare diseases. OGT has utilised Emory’s extensive expertise to identify the most up-to-date gene content and subsequently research-validate the array performance, maximising the likelihood of detecting causative variation.

Dr Lewis and Dr Farrow both employ the CytoSure MREA and find that, combined with targeted exome NGS, this provides a cost-effective workflow for comprehensive mutational analysis. Dr Farrow comments, “NGS is currently limited in its detection of CNVs, and we employ the MREA alongside both genome and exome sequencing for detecting additional variants, strengthening the evidence towards the detection of a particular disorder.” She went on to explain, “The MREA has been used in our research looking at families where NGS has identified one compelling variant, and we are looking for that second disease-linked variant. We see great benefits for this approach in instances where there is no clear phenotype associated with a particular disorder.”
Despite the evolving capabilities of NGS, the current importance of the microarray is highlighted by Dr Lewis, “It may take many years before NGS performance and analysis is optimised to allow detection of CNVs directly from the NGS data. Until such time, we will continue to employ the MREA.”
The content of the microarray determines its performance, and for high-resolution CNV detection with a high signal-to-noise ratio, many aspects must be carefully considered, and the preliminary content must also undergo careful validation. The combination of expertise from OGT and Emory working on the pre-optimisation and validation has led to the MREA delivering the most insightful coverage with a high signal-to-noise ratio.
Researchers can be confident that the existing gene content and probes will perform to the highest standards, as Dr Lewis adds, “One of the main reasons why we chose the OGT MREA was due to the trusted consortium of Emory Genetics Laboratory working with OGT throughout the design process.” This process is explained in more detail below.
1. Ensuring medically-relevant coverage
Defining the microarray content essentially starts when a published paper shows a particular gene is linked with a disorder. Further investigations subsequently focus on determining the exons involved, the mutation spectrum and designing the clinical tests.
Professor Madhuri Hegde and her team at Emory Genetics Laboratory run a monthly internet database search for identifying newly discovered medically-relevant genes. Through further reading they decide if the gene is significant, and from this a list is curated. A continuing open dialogue between Emory and OGT ensures the gene content of CytoSure arrays reflects the latest research findings, allowing scientists to keep up to date with the latest medical research.
2. Probe design
For the MREA, the probes are designed by the OGT bioinformatics team, utilising proprietary design algorithms to ensure optimal probe performance. The probes are highly sensitive, specific and they are also designed to anneal under similar conditions to ensure uniform hybridisation.
3. Validation
Validation work by Emory utilising clinical research samples ensures sufficient coverage and optimal performance of the MREA. This includes, for example, comparing results to that of a disease-specific targeted array (Figure 1). Comparison with this smaller format design, which includes a higher density of probes for the disease-relevant region, verifies the coverage of the MREA is sufficient to detect the same aberrations. The coverage and content is modified until the performance meets Emory’s exacting specifications.
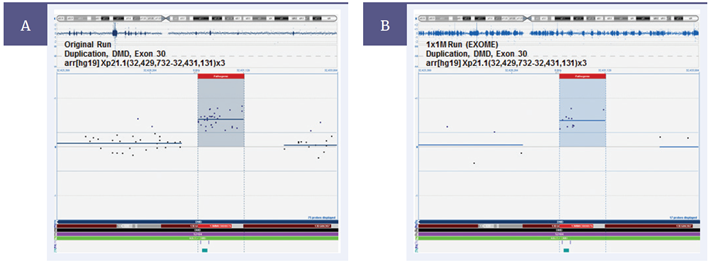 Figure 1: Validating the CytoSure Medical Research Exome Array. As part of the validation process, Emory Genetics Laboratory compared CNV detection between the OGT CytoSure Molecular Array for Duchenne Muscular Dystrophy (DMD) (8x60k) and the CytoSure MREA. (A) The DMD array detects the duplication of Exon 30. (B) This result is duplicated with the MREA, validating the detection of this CNV. [Image courtesy of Professor M. Hegde, Emory Genetics Laboratory].
Figure 1: Validating the CytoSure Medical Research Exome Array. As part of the validation process, Emory Genetics Laboratory compared CNV detection between the OGT CytoSure Molecular Array for Duchenne Muscular Dystrophy (DMD) (8x60k) and the CytoSure MREA. (A) The DMD array detects the duplication of Exon 30. (B) This result is duplicated with the MREA, validating the detection of this CNV. [Image courtesy of Professor M. Hegde, Emory Genetics Laboratory].
The ability to customise OGT’s MREA, enables researchers to utilise this single validated platform as a base for a host of more specific studies. From the MREA, Dr Lewis has created customised arrays for a number of individual projects:
Although the Alport’s panel contains only three genes (COL4A5, COL4A3, and COL4A4), MLPA kits are only available for two of the genes, as Dr Lewis explains: “Neither of the MLPA kits interrogate all exons of the targeted gene, and the only means to achieving comprehensive analysis for Alport’s syndrome is through targeted sequencing and microarray analysis.”
She also modified the design of custom arrays using MREA probes. “We first compared the MREA against our internally validated list of genes. We identified exons with fewer than four probes where we requested additional coverage, and for those novel genes of interest not yet included on the medical exome.” For example, the group’s microarray data recently uncovered the existence of a processed pseudogene for SMAD4.1 “To aid in distinguishing the presence of a processed pseudogene from a true gene duplication, we required additional probes in the intronic regions of SMAD4,” says Dr Lewis. “OGT were quick to respond to our requests with the design of additional probes, and the ability to work closely with the OGT design team to optimise array content is a great advantage for us.” The MREA also contains additional space. “We add backbone probes or additional targeted probes. However, we keep 5–10% free for future additions to the design.”
While many array platforms carry high numbers of probes, the key to a higher resolution is the number of probes in the specific region of interest. Exon-focused designs such as that of the MREA enable high resolution coverage down to 12bp in the case of a deletion in intron 4 of the DBT gene.2 Drs Lewis and Farrow both find this resolution highly advantageous. “We have validated a variety of CNV sizes, but we are particularly interested in detection at the level of the single exon,” says Dr Lewis. Dr Farrow comments: “The OGT MREA provides us with the exon-level resolution needed to detect CNVs over the medical exome that are missed by NGS and traditional microarray designs, providing additional insights into the mutation spectrum of the sample.”
Medical researchers, such as Dr Lewis and Dr Farrow, employ a suite of available technologies to provide comprehensive analysis of genetic disorders, guiding genetic research as well as driving the applied therapeutic strategies of the future. “Even as NGS continues to develop, microarrays remain critical to detecting CNVs,” says Dr Farrow.
Emory Genetics Laboratory and OGT have worked side-by-side to develop the OGT CytoSure Medical Research Exome array, defining the content based on direct clinical experience. The medical genetics research community now has access to a fully research-validated microarray, optimised for high-performance CNV detection in medically-relevant genes, which can even be customised for specific studies. The MREA content is continually updated through the ongoing partnership with OGT and Emory, expanding the medical exome with those genomic regions that are becoming increasingly recognised in disease.
CytoSure®: For Research Use Only; Not for Diagnostic Procedures.
