Colorectal cancer (CRC) is the third most common cancer in men (746,000 cases, 10.0% of the total and the second in women (614,000 cases, 9.2% of the total) worldwide1. Next generation sequencing (NGS) has enabled the simultaneous study of mutations in high-penetrance colorectal cancer genes. These include KRAS, APC and TP53 as well as more moderate-risk genes such as ERBB2, PTEN and BRAF2.
Choose your ideal colorectal cancer NGS panel from our range of fully tested and optimised NGS panel content. Simply mix and match the genes or individual exons you require to get the most out of your sequencing runs. Use in conjunction with the SureSeq™ FFPE DNA Repair Mix* for improved NGS library yields, %OTR and mean target coverage from challenging FFPE derived samples.
Getting started with your SureSeq myPanel NGS Custom Panel could not be simpler, find out more in this video…
* Exon examples not yet available
We have a regularly updated, expert-curated library of pre-optimised cancer panel content for you to select from. Simply mix and match the gene, exonic or intronic content you need to create a colorectal cancer NGS cancer panel that meets your exact requirements.
KRAS mutations are found in approximately 35-45% of colorectal cancers with around 80% occurring in codon 12 and 15% in codon 13 of exon 2; other commonly reported mutations are found in exons 3 and 43. The tumor suppressor gene APC plays an important role in CRC development. Absence of the APC protein leads to accumulation of betacatenin in the cytoplasm, which may contribute to tumour progression. 60% of all somatic mutations in APC occur within the mutation cluster region between codons 1286 and 1513 on exon 154. Figures 1 and 2 illustrate the superior uniformity of coverage of these key genomic regions.
Approximately 8-15% of colorectal cancers involve mutations in the BRAF gene, with up to 90% of these a result of a mutation at V600E, located on exon 155. In TP53, another frequently mutated cancer gene, point mutations are predominantly located in exons 5-82, however sequencing is often hampered by the GC-rich content, which can lead to technical challenges in assay design and analysis. OGT’s innovative bait design overcomes this issue, offering a high level of uniform coverage for these difficult genes to sequence in FFPE samples (Figures 3 and 4).
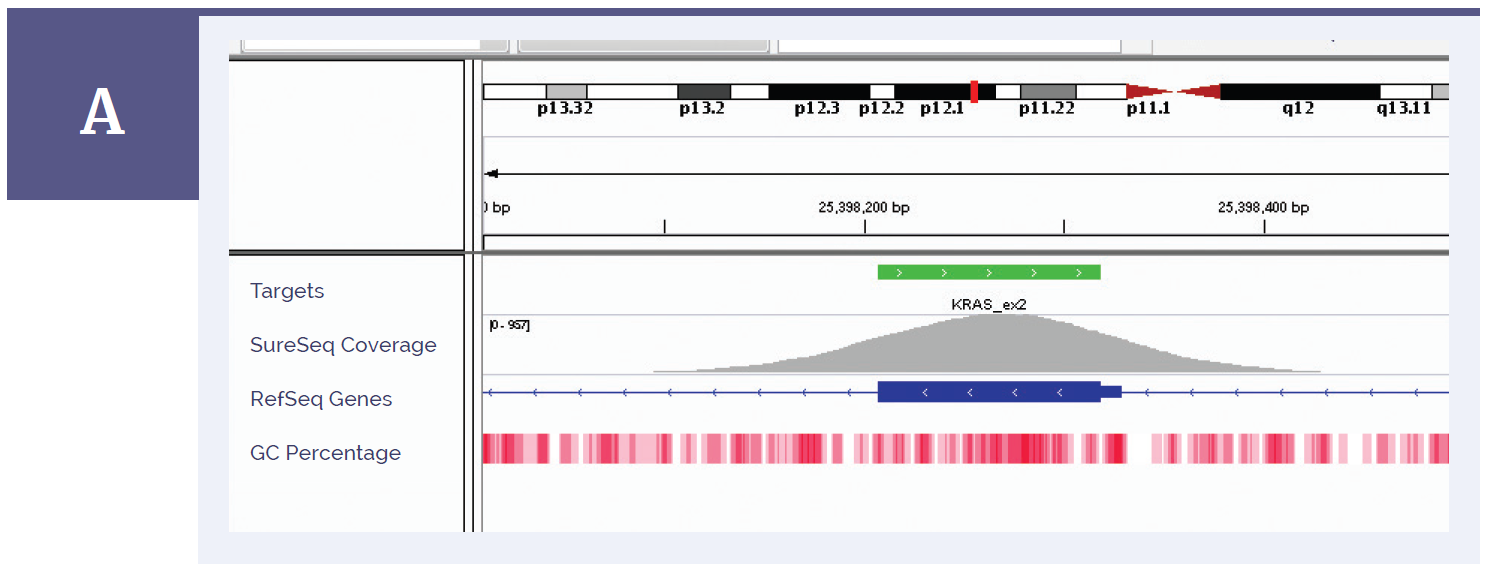
Figure 1a: KRAS exon 2. Depth of coverage per base (grey). Targeted region (green). Gene coding region as defined by RefSeq (blue). GC percentage (red).

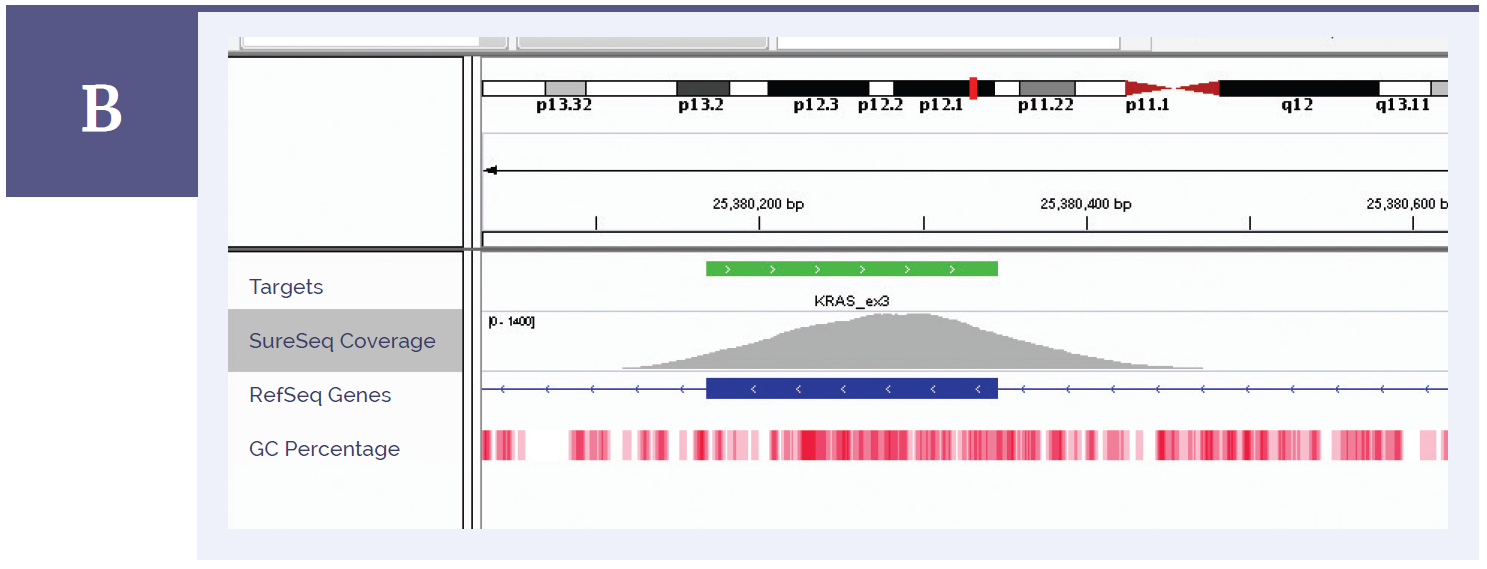
Figure 1b: KRAS exon 3. Depth of coverage per base (grey). Targeted region (green). Gene coding region as defined by RefSeq (blue). GC percentage (red).

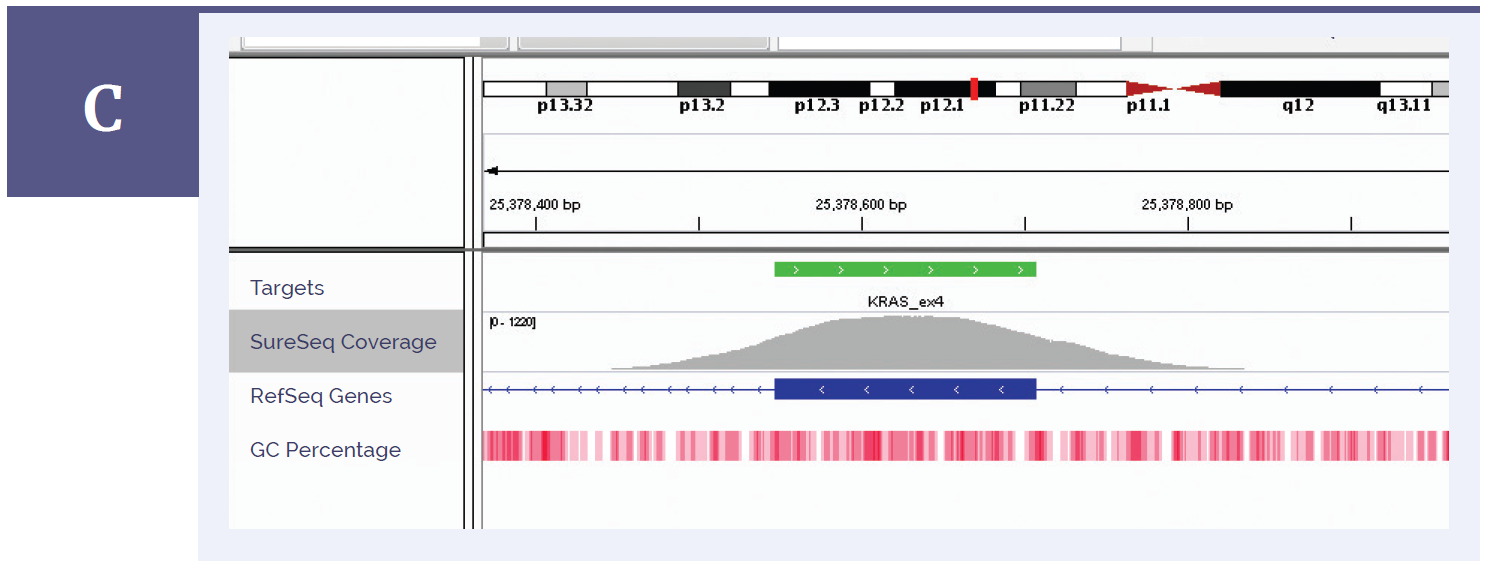
Figure 1c: KRAS exon 4. Depth of coverage per base (grey). Targeted region (green). Gene coding region as defined by RefSeq (blue). GC percentage (red).

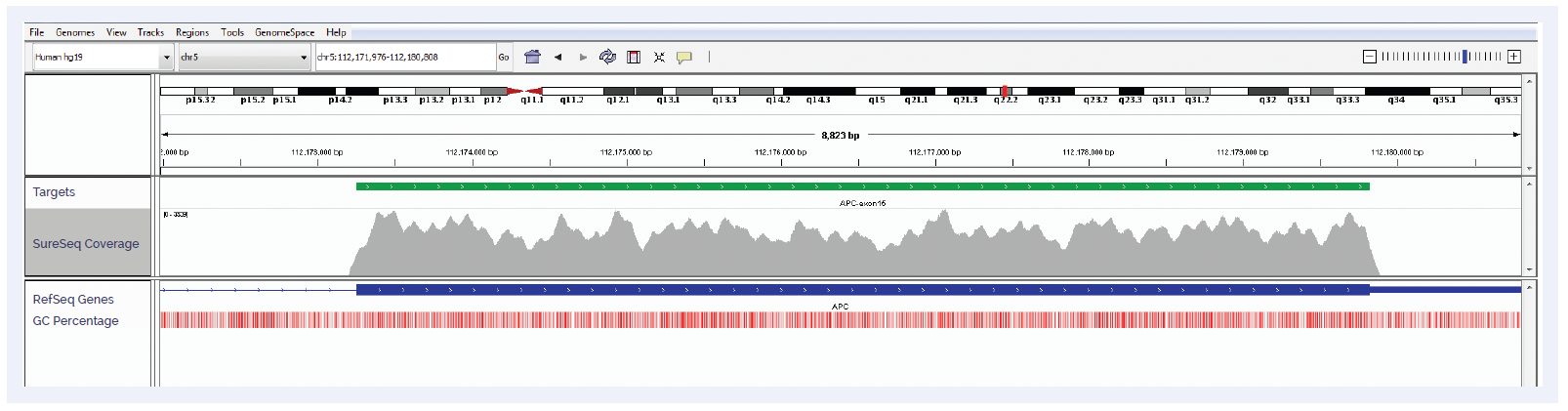
Figure 2: APC exon 15 coverage. Depth of coverage per base (grey). Targeted region (green). Gene coding region as defined by RefSeq (blue). GC percentage (red).


*The SureSeq FFPE DNA Repair Mix can only be purchased in conjunction with SureSeq NGS panels, not as a standalone product.


