Want to download this as a PDF? Download now
Jacqueline Chan, Juliette Forster, William Wright, Graham Speight
To overcome these issues, we have used the SureSeq™ FFPE DNA Repair Mix*, in combination with a hybridisation-based NGS custom enrichment panel, the SureSeq Ovarian Cancer Panel (Figure 1) to identify somatic variation in key DNA repair genes associated with ovarian cancer.
![]() Table 1: Key ovarian cancer-related genes in the SureSeq Ovarian Cancer Panel.
Table 1: Key ovarian cancer-related genes in the SureSeq Ovarian Cancer Panel.
To evaluate the application of a hybridisation-based approach we:
To confidently call low level variants, NGS reads need to be evenly distributed across all regions of interest. Uniformity of coverage is a useful value with which to compare this distribution and can be expressed as the percentage of target bases that have greater than 20% of the mean coverage. As reported extensively in the literature1-3, the uniformity of coverage from capture-based approaches such as SureSeq consistently outperform those enriched using amplicon-based methods (Figure 2). Furthermore, in our sample set we found the high levels of uniformity are maintained when starting with ~250 ng DNA (light blue bars).
The uniformity of the coverage for most samples is greater than 99% of bases covered at 20% of the mean, ensuring that all bases within the panel can be assessed.
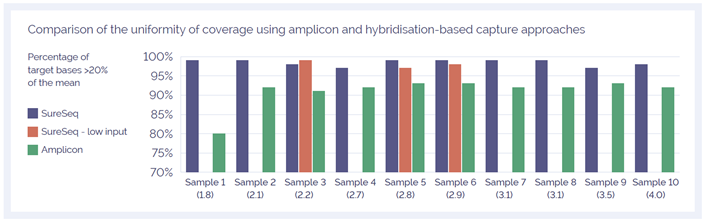 Figure 2: Assessment of the uniformity of sequencing coverage from FFPE-derived DNA using an amplicon and the SureSeq hybridisation capture-based approaches. Enrichment by SureSeq sequence capture (dark blue bars) demonstrates better uniformity than that of an amplicon-based approach (green bars). The level of uniformity is maintained when starting with ~250 ng DNA (light blue bars). Samples are ordered by increasing DNA Integrity Number (DIN) determined by Agilent 2200 TapeStation - value in brackets.
Figure 2: Assessment of the uniformity of sequencing coverage from FFPE-derived DNA using an amplicon and the SureSeq hybridisation capture-based approaches. Enrichment by SureSeq sequence capture (dark blue bars) demonstrates better uniformity than that of an amplicon-based approach (green bars). The level of uniformity is maintained when starting with ~250 ng DNA (light blue bars). Samples are ordered by increasing DNA Integrity Number (DIN) determined by Agilent 2200 TapeStation - value in brackets.
The superior uniformity of coverage enables reliable identification of somatic single nucleotide variants (SNVs) and indels in solid tumour samples. Figure 3 illustrates some examples of somatic deletions (panel A) and SNVs (panel B) that have been found in exon 6 of TP53 from FFPE blocks of type II EOC samples.
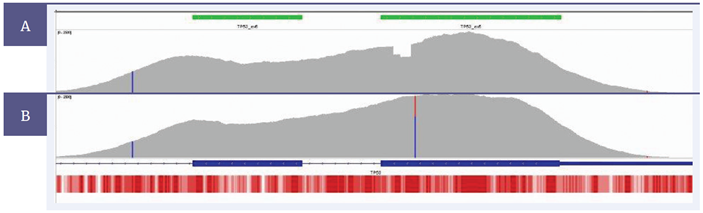 Figure 3: Sequence coverage of TP53 exons 5 and 6 from type II EOC FFPE-derived DNA. The SureSeq hybridisation-based capture approach achieved a high depth of coverage over the GC rich exon 5 of TP53. This has enabled a 18 bp deletion - panel [A], and a C->T SNV (R175H) - panel [B] to be detected at a minor allele frequency of 23.5% and 33.3%, respectively. Targeted region - green; depth of coverage per base - grey; gene coding region as defined by RefSeq - blue; GC percentage- red; visualised using Integrated Genomics Viewer4.
Figure 3: Sequence coverage of TP53 exons 5 and 6 from type II EOC FFPE-derived DNA. The SureSeq hybridisation-based capture approach achieved a high depth of coverage over the GC rich exon 5 of TP53. This has enabled a 18 bp deletion - panel [A], and a C->T SNV (R175H) - panel [B] to be detected at a minor allele frequency of 23.5% and 33.3%, respectively. Targeted region - green; depth of coverage per base - grey; gene coding region as defined by RefSeq - blue; GC percentage- red; visualised using Integrated Genomics Viewer4.
We found pre-treatment with the SureSeq FFPE DNA Repair Mix improves the mean target coverage of formalin comprised samples (Horizon Diagnostics), thereby increasing the flexibility of the assay (Figure 4). Use of the repair mix also enables a reduced DNA input down to 50 ng whilst maintaining a good depth of coverage (Figure 5).
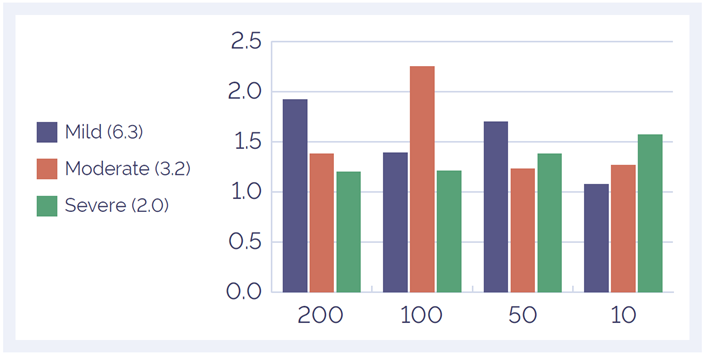 Figure 4: Improvement in mean target coverage through use of the FFPE repair mix. All three standards showed an improvement in coverage of the target bases when treated with FFPE repair mix. The mean fold increase across all sample types and input amounts was 1.5x. Samples are ordered by DNA Integrity Number (DIN) - value in brackets.
Figure 4: Improvement in mean target coverage through use of the FFPE repair mix. All three standards showed an improvement in coverage of the target bases when treated with FFPE repair mix. The mean fold increase across all sample types and input amounts was 1.5x. Samples are ordered by DNA Integrity Number (DIN) - value in brackets.
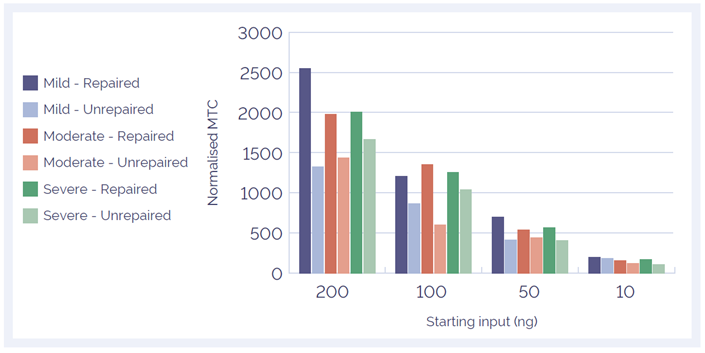 Figure 5: Effect of reduced amount of DNA input on mean target coverage. The mean target coverage decreases with reduced amount of input DNA but the FFPE repair mix treatment helps maintain a good depth of coverage.
Figure 5: Effect of reduced amount of DNA input on mean target coverage. The mean target coverage decreases with reduced amount of input DNA but the FFPE repair mix treatment helps maintain a good depth of coverage.
All samples had 100% concordance for 20 reported variants with 97.5% having allele frequencies within 5% of the expected value (Table 1).
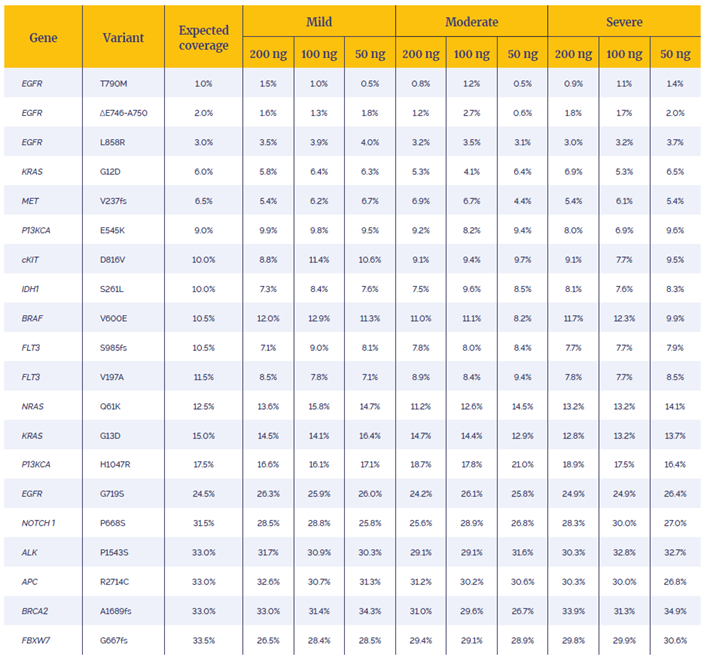 Table 1: Difference between the expected and observed allele frequency in a characterised sample. The variants - 15 SNVs and 5 deletions, ranging from 1% to 33.5% minor allele frequency, were determined using OGT’s Interpret software.
Table 1: Difference between the expected and observed allele frequency in a characterised sample. The variants - 15 SNVs and 5 deletions, ranging from 1% to 33.5% minor allele frequency, were determined using OGT’s Interpret software.
We have shown that the use of SureSeq hybridisation based panels with the SureSeq FFPE repair mix provides:
Research samples provided by -
aProf. Charlie Gourley (Cancer Research UK Edinburgh Centre)
bProf. Robert Zelllinger and Dr. Nicole Concin (Medical University of Vienna and Medical University, Dept. of Gynecology and Obstetrics, Vienna, Austria)
*The SureSeq FFPE DNA Repair Mix can only be purchased in conjunction with SureSeq NGS panels, not as a standalone product.
SureSeq™: For Research Use Only; Not for Diagnostic Procedures.
