Want to download this as a PDF? Download now
 Even when considering the emergence of next generation sequencing technologies, array comparative genomic hybridisation (aCGH) remains a staple of the modern clinical genetics research laboratory, detecting with ease and confidence the genetic aberrations linked to many developmental disorders.
Even when considering the emergence of next generation sequencing technologies, array comparative genomic hybridisation (aCGH) remains a staple of the modern clinical genetics research laboratory, detecting with ease and confidence the genetic aberrations linked to many developmental disorders.
The astonishing pace of genetic discoveries means that novel disease-relevant variants are frequently coming to light in the scientific literature. Microarrays must therefore be routinely updated, as published genetic discoveries guide the content focus towards the most insightful regions. One such microarray is the CytoSure® Constitutional v3 platform from Oxford Gene Technology (OGT), which provides enhanced exon-level coverage of developmental disorder genes1. This is based on a combination of the most up-to-date and relevant content from the recent Deciphering Developmental Disorders (DDD) study2 and latest updates from ClinGen*, the Clinical Genome Resource3. The highly disease-relevant content combined with single-exon copy number variation (CNV) resolution is a key reason why many genetics laboratories are choosing to utilise this platform — including the Constitutional Cytogenetics laboratory, University Hospitals Leuven (Belgium).
An important factor when considering the capabilities of microarray technology is the data quality, and it is vital that scientists have absolute confidence when reporting a variant. As such, quality control is of utmost importance at the Constitutional Cytogenetics laboratory, which actively participates in the Cytogenetic External Quality Assessment Service (CEQAS) programme for ensuring optimum quality standards. The laboratory also forms part of the Belgian Society for Human Genetics (BeSHG) Workgroup on Constitutional Post- and Prenatal (Molecular) Cytogenetics, supporting the optimisation of working guidelines in Belgium and across Europe.
As part of the Centre for Human Genetics, the Constitutional Cytogenetics laboratory, led by Professor Joris Vermeesch, has been using OGT’s CytoSure microarrays since 2009, and have adopted the CytoSure Constitutional v3 array for prenatal and postnatal genetic research. As part of implementing the new array platform, thorough validation ensures results meet the high quality standards required. Interestingly, following a comprehensive validation of OGT’s Constitutional v3 array, significant advantages have also been revealed. Laboratory manager and quality assessor at the Constitutional Cytogenetics laboratory, Dr Kris Van Den Bogaert, discusses these advantages and the process of implementing the new CytoSure Constitutional v3 array.
The Centre for Human Genetics at Leuven University is amongst the largest in Europe, with the cytogenetics laboratory processing approximately 15,000 samples per year. The centre performs preimplantation, invasive and non-invasive prenatal, as well as postnatal genomic analysis. Particularly for the prenatal and postnatal samples, the microarray forms a key element in furthering the understanding of genetic disorders (Figure 1).
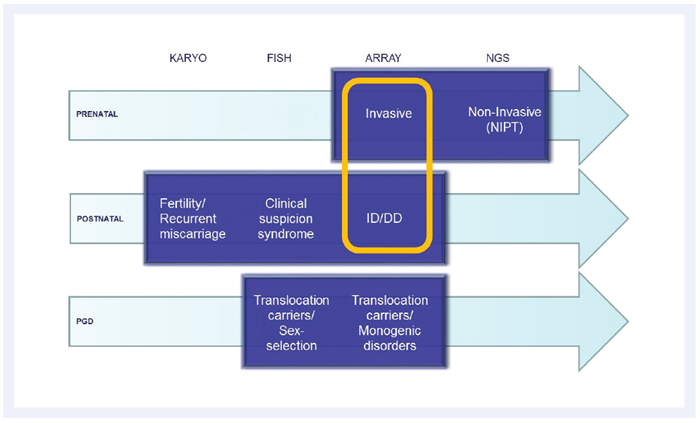 Figure 1: Genetic analysis at Leuven. Of the four main techniques, the microarray is utilised to investigate prenatal invasive samples and postnatal samples where intellectual disability/developmental disorders (ID/DD) are suspected.
Figure 1: Genetic analysis at Leuven. Of the four main techniques, the microarray is utilised to investigate prenatal invasive samples and postnatal samples where intellectual disability/developmental disorders (ID/DD) are suspected.
Investigating the underlying causes of intellectual disability and developmental disorders of unknown origin, the microarray is employed to detect deletions and duplications across the entire genome in postnatal samples. Invasive prenatal samples taken from amniocentesis or chorionic villi are also investigated using microarray. From 2013, the national consensus in Belgium was to use microarray for all invasive prenatal samples, instead of conventional karyotyping. Kris explains: “We use an 8x60k array format for all invasive prenatal samples, which is a lower resolution than our postnatal 4x180k array. The lower resolution is due to a national consensus4, which is a balanced decision between maximising the detection yield of the test and minimising the number of variants of unknown significance.”
If no duplications or deletions are detected from microarray analysis, postnatal samples are subsequently often investigated with NGS. Although the accessibility of NGS for the clinical research laboratory is increasing, the bioinformatics is not yet optimised to detect CNV at the resolution, or with the reliability, afforded. by the microarray. Interestingly, Kris highlights another synergy between these two technologies, with the microarray confirming NGS results: “For our colleagues working with NGS, in a few cases their results have pointed to a possible exonic deletion or duplication, and they have asked whether we can confirm this using the CytoSure Constitutional v3 platform. It has also been the case that NGS clearly finds a variant potentially underlying an associated autosomal recessive disorder. We then search for a deletion or duplication to explain the cause of the condition.”
The laboratory developed and implemented the 1Mb BAC array in 2004. The printing of arrays was both labour intensive and time consuming, with only a basic in-house developed analysis toolkit and low resolution. In 2009 the laboratory switched to the OGT CytoSure platform. Kris comments: “Moving to the OGT array platform was decided primarily due to the interface of the CytoSure Interpret Software supplied alongside the arrays, which is very user-friendly and customisable. It also includes data tracks that can be adapted to include the information we need for interpretation of the variants.”
The laboratory relied on OGT’s support to optimise their complete workflow, especially when implementing a proprietary experimental design for the postnatal microarrays. “Our experimental design has very specific requirements, and OGT was of great help in adapting the software to enable the analysis of our samples, proving a significant advantage for our workflow”. It was also possible to connect OGT’s CytoSure Interpret Software with third party software, including data storage programmes to allow the easy export of data, as Kris explains: “Interaction with our in-house sample database means we can easily import and export data from CytoSure Interpret Software, including automatically importing sample identification details, making life much easier.”
Transitioning to OGT’s Constitutional v3 microarray was decided for several reasons, as Kris comments: “The new arrays provide us with enhanced exon-level coverage for specific developmental disorder genes. It is of great value to us and clinical research as a whole that the selection of the genes on the array is based on the most up-to-date findings.” Before implementing the array, the laboratory performed a comprehensive validation process.
Validation method:
For both 4x180k and 8x60k arrays (utilised for post- and prenatal research, respectively), prospective validation was performed by parallel processing of samples on OGT’s CytoSure ISCA v2 and CytoSure Constitutional v3 microarray platforms. The QC metrics that were compared included Derivative Log Ratio (DLR) spread and signal-to-noise ratio. While the DLR spread is perhaps the most important QC metric and calculates the probe-to-probe log ratio noise, the signal-to-noise ratio indicates how clearly the signals can be detected above the background level. A poor DLR spread (>0.3) or signal-to-noise ratio (<30) will mean that it is more difficult to accurately call a variant. In addition to QC metrics, the analytical accuracy as defined by concordance between the ISCA v2 and Constitutional v3 platforms was also assessed. Sample numbers were n=80 and n=41, respectively for the 4x180k and 8x60k arrays. Retrospective validation was also performed, with samples carrying exonic deletions/duplications in the NF1 and BRCA2 genes first confirmed by Multiplex Ligation-dependent Probe Amplification (MLPA). These samples (n=4) were then tested on the Constitutional 4x180k v3 array.
Validation results:
For the prospective validation, both Constitutional v3 array formats, 4x180k and 8x60k, were compared to their respective ISCA v2 array design. DLR was slightly improved and the signal-to-noise slightly increased on the Constitutional v3 4x180k microarray (Figure 2), and were similar on the Constitutional v3 8x60k array (Figure 3). The variance was small and results from all metrics with all arrays was significantly above minimum expected levels indicating the high data quality seen with the ISCA v2 had been maintained on the Constitutional v3 design.
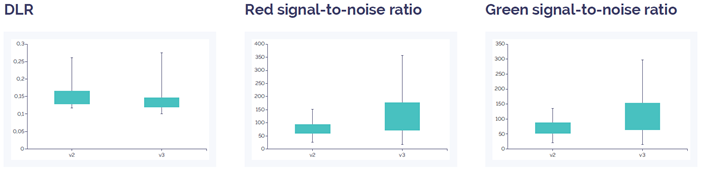 Figure 2: CytoSure Constitutional v3 4x180k array data quality compared to CytoSure ISCA v2 4x180k. DLR is slightly improved in the v3 array compared to the v2 array, and the signal-to-noise ratio is increased for both red and green. Data quality for both arrays is well above quality threshold (DLR spread <0.3; signal-to-noise ratio >30).
Figure 2: CytoSure Constitutional v3 4x180k array data quality compared to CytoSure ISCA v2 4x180k. DLR is slightly improved in the v3 array compared to the v2 array, and the signal-to-noise ratio is increased for both red and green. Data quality for both arrays is well above quality threshold (DLR spread <0.3; signal-to-noise ratio >30).
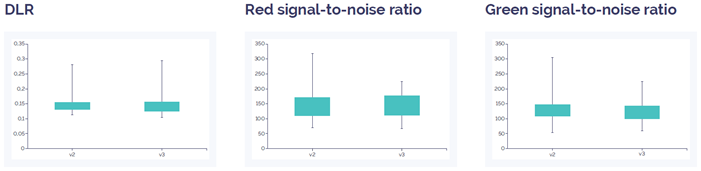 Figure 3: CytoSure Constitutional v3 8x60k array data quality compared to CytoSure ISCA v2 8x60k. DLR spread and signal-to-noise level show an equivalent value. Overall this indicates a similar level of quality between the two array platforms. Data quality for both arrays is well above quality threshold (DLR spread <0.3; signal-to-noise ratio >30).
Figure 3: CytoSure Constitutional v3 8x60k array data quality compared to CytoSure ISCA v2 8x60k. DLR spread and signal-to-noise level show an equivalent value. Overall this indicates a similar level of quality between the two array platforms. Data quality for both arrays is well above quality threshold (DLR spread <0.3; signal-to-noise ratio >30).
While all of the 41 samples for the 8x60k array exhibited complete concordance, for the 4x180k array, 77 out of the 80 samples were concordant (see examples in Figure 4). This included two aberrations detected only with the Constitutional v3 microarray, pointing to its increased coverage (Figure 5). It is important to note that the three samples were small variants below 30kb, and therefore below the 200kb cut-off used for standard reporting. Unless aberrations below 200kb are directly linked with a specific phenotype, this lack of concordance would not impact the final report. However, the enhanced coverage of the Constitutional v3 microarray enables high-resolution exon-level CNV detection, and Kris explains how such smaller single-exon micro-deletion/ micro-duplications are in fact coming to light as increasingly significant: “In the research setting, NGS data is also detecting these smaller aberrations, and evidence is amounting to show that exon-level coverage and detection is important for understanding the underlying genetic aberrations of a certain phenotype.”
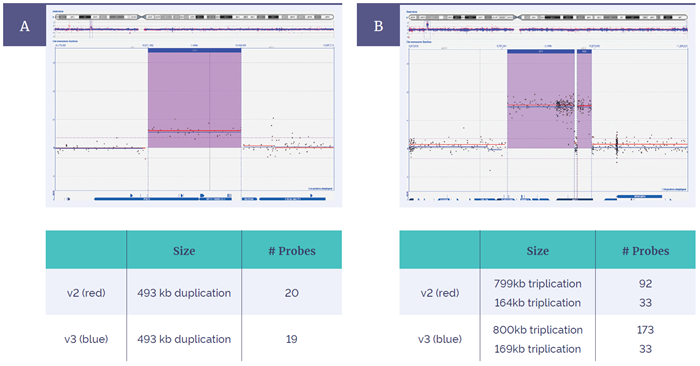 Figure 4: Concordance between the CytoSure ISCA v2 4x180k and Constitutional v3 4x180k. Within CytoSure Interpret Software, aberrations are intuitively shown in context of chromosomal location (above), and gene content (below). Both arrays detected (A) a 493kb duplication and (B) two flanking triplications on chromosome X, with more probes added in this area on the v3 platform.
Figure 4: Concordance between the CytoSure ISCA v2 4x180k and Constitutional v3 4x180k. Within CytoSure Interpret Software, aberrations are intuitively shown in context of chromosomal location (above), and gene content (below). Both arrays detected (A) a 493kb duplication and (B) two flanking triplications on chromosome X, with more probes added in this area on the v3 platform.
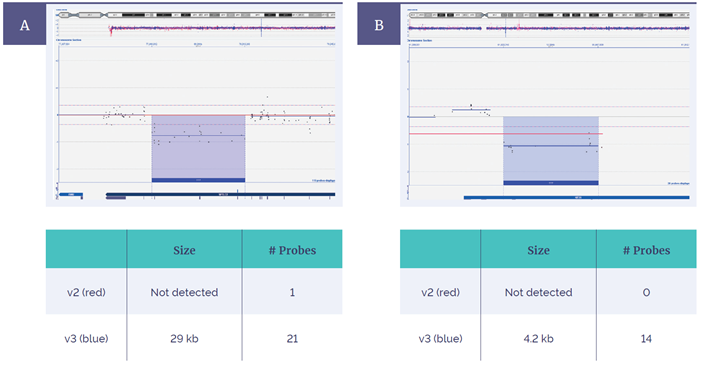 Figure 5: Extended coverage with the CytoSure Constitutional v3 4x180k array compared to the CytoSure ISCA v2 microarray 4x180k. In 2 of 80 samples, an increased exonic probe coverage resulted in enhanced aberration detection of a 29kb deletion within the SPTLC2 gene (A) and a 4.2kb deletion within the KIF2A gene (B).
Figure 5: Extended coverage with the CytoSure Constitutional v3 4x180k array compared to the CytoSure ISCA v2 microarray 4x180k. In 2 of 80 samples, an increased exonic probe coverage resulted in enhanced aberration detection of a 29kb deletion within the SPTLC2 gene (A) and a 4.2kb deletion within the KIF2A gene (B).
To support the accuracy and therefore confidence of the microarray within the clinical research laboratory, retrospective validation was also performed. Samples that were first confirmed with MLPA for exonic copy number variations in the BRCA2 and NF1 gene (Figure 6) subsequently found complete concordance. Moreover, coverage was significantly enhanced for both genes on the Constitutional v3 compared to the ISCA v2, as Kris explains: “On the v2 platform we would never be able to detect these aberrations. We wanted to check the v3 was sensitive enough to detect these small aberrations — and it was.”
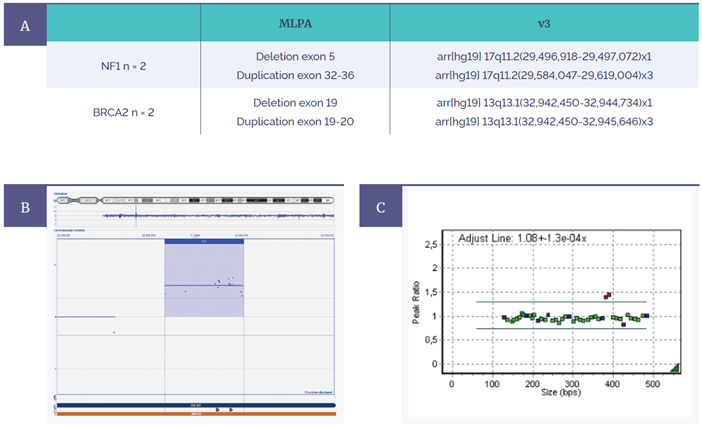 Figure 6: Retrospective analysis shows complete concordance between MLPA and the CytoSure Constitutional v3 4x180k microarray. (A) Four variants were tested for in two genes (NF1 and BRCA2). (B) Constitutional v3 microarray data from BRCA2 duplication of exon 19-20. (C) Corresponding MLPA data showing duplicated exons in red.
Figure 6: Retrospective analysis shows complete concordance between MLPA and the CytoSure Constitutional v3 4x180k microarray. (A) Four variants were tested for in two genes (NF1 and BRCA2). (B) Constitutional v3 microarray data from BRCA2 duplication of exon 19-20. (C) Corresponding MLPA data showing duplicated exons in red.
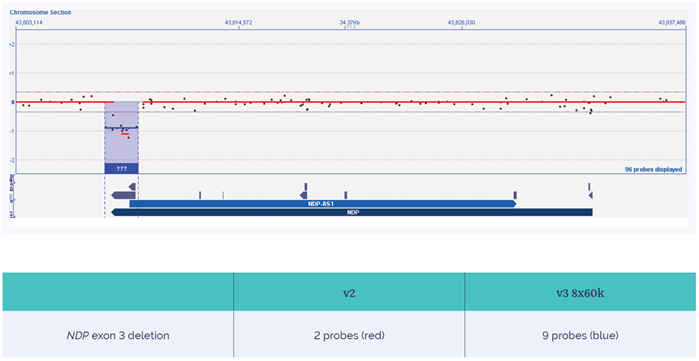 Figure 7: Increased yield of a family-specific exon 3 deletion of the NDP gene on the CytoSure Constitutional v3 microarray 4x180k. The NDP exon 3 deletion is detected with higher confidence using OGT’s CytoSure Constitutional v3 array, which has nine probes in the 4x180k format, compared to the two probes of the 4x180k ISCA v2 array.
Figure 7: Increased yield of a family-specific exon 3 deletion of the NDP gene on the CytoSure Constitutional v3 microarray 4x180k. The NDP exon 3 deletion is detected with higher confidence using OGT’s CytoSure Constitutional v3 array, which has nine probes in the 4x180k format, compared to the two probes of the 4x180k ISCA v2 array.
Although more data is currently being generated to confirm the improved detection levels enabled by the new array, Kris comments on the validation success: “From the samples in the validation cohort, we expect to observe an increase in the level of disease-relevant variants detected. However, we will only be able to provide numbers once we start using this daily, running hundreds and thousands of samples.” Through advancing clinical research, this improvement will have significant implications in the healthcare setting. An example from the laboratory involved detection of a family-specific deletion of exon 3 in the NDP gene due to the increased exonic probe coverage.
Compared to the two probes of the ISCA v2, the Constitutional v3 array includes nine probes on the 4x180k format located in this specific exon 3 deletion. Confidence increases with probe density across this region, with the Constitutional v3 array providing the confidence necessary to call this small, exonic aberration (Figure 7).
In summary, the validation of OGT’s CytoSure Constitutional v3 microarray at the Constitutional Cytogenetics laboratory of Leuven University has been a core part of its implementation. The study indicated equivalent QC metrics between the new platform and the platform that the laboratory already had in place. Above the variant reporting cut-off of 200kb, the two platforms also exhibited full concordance in prospective analysis, while retrospective analysis of aberrations within the NF1 and BRCA2 genes also exhibited complete concordance between MLPA-confirmed samples and the CytoSure Constitutional v3 array.
Most importantly, the new platform has increased coverage for clinically-relevant regions which results in the detection of more disease relevant variants. During validation there have been instances where the new Constitutional v3 array was able to call small aberrations that would have been missed on the ISCA v2 array, e.g. an exonic deletion within NDP. As Kris confirms: “It is this extra coverage that leads to understanding the aberrations underlying a growing number of disorders, and this is a real added value of the v3 platform.” Following the validation process and the positive results acquired, the laboratory is now moving to the CytoSure Constitutional v3 4x180k array in the postnatal setting, and is also set to finalise the validation for the prenatal investigations with the CytoSure Constitutional v3 8x60k array.
* Formerly known as ISCA/ICCG.
CytoSure®: For research use only; not for use in diagnostic procedures.
